OrganismView
OrganismView is CoGe's tool for searching for the genome of an organism of interest, and getting an overview of genomic information
Contents
Introduction
CoGe is designed to store multiple versions of any genome from multiple organisms from all domains of life in any state of assembly and annotation. This includes bacteria, archaea, eukaryotes, organelles, viruses, and sub-genomes such as plasmids. The genomic sequence can also exist in different states such as being partially assembled, fully assembled, completely unmasked, masked for repeats, etc. Also, there can exist different sets of genomic features and annotations that. OrganismView allows users to get detailed information about the genomes available for a given organism, and provides links to other tools in CoGe to extract and visualize various types of genomic information.
Getting Started
Most organisms in CoGe use the scientific binomen (i.e. Genus species; e.g. Escherichia coli) for their name and full Linnaean lineage for their description (e.g. Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Escherichia).
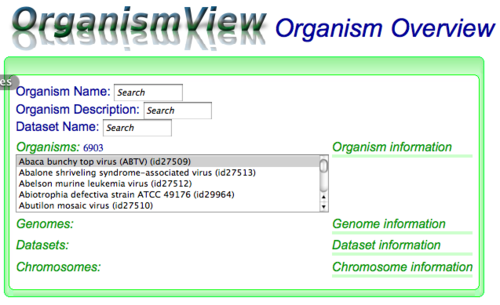
To search for an organism, type any part of their name or description in OrganismView's "Organism Name" or "Organism Description" search box respectively. OrganismView will start searching for anything that matches and displays those organisms in a selectable list below the header "Organisms:". The small number next to the "Organisms:" header is the count of the number of organisms whose name or description matched your search term. Next, just scroll through the list and select your organism. Information about it will start to automatically appear in the other sections of OrganismView.
Organization and Information
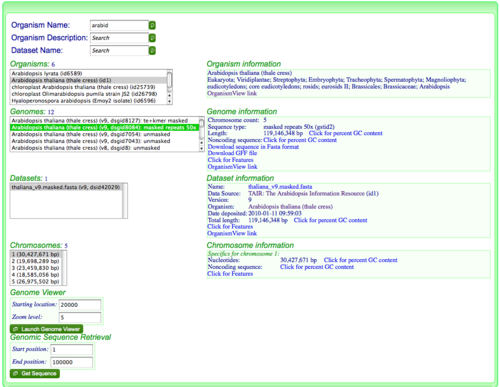
When an organism is selected, various types of information are shown in varying degrees of scope (listed largest to smallest):- Organism: top level list of organisms
- Genome:whole genome information. For those interested in CoGe's database, this refers to the Dataset group table.
- Dataset: a given genome is comprised of one or more datasets. Different genomic resources organism genomic information differently and this allows for a representation of how an organism's genome was acquired. For example, each chromosome may come from a separate data file.
- Chromosome: the list of chromosomes for a selected dataset.
OrganismView is organized such that the above information is listed from the top to the bottom of the screen. Each scope level is organized such that selectable lists for the scope is shown on the left of the screen, and information about the selection is shown to the right.
Organism Information
Shows the name and description for the selected organism.
Genome Information
Overview of the genome:
- Chromosome count (will be very high for partially assembled genomes)
- Sequence type: Unmasked sequence, masked sequence
- Total length: For all datasets making up this genome which may include plasmids, organelles, etc. depending on how the "genome" was defined by whomever sequenced the genome. This will automatically calculate the percent GC for genomes smaller than 10 megabases, otherwise the user can click on a link to calculate percent GC content.
- Non-coding sequence: A link that will calculate the length and GC content of non-protein coding sequence
Download Data
Fasta Sequences
Use this link to download the entire genome's sequence
Export GFF
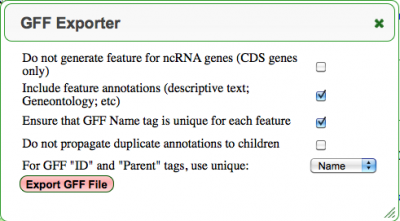
Use this link to open the GFF Exporter Dialog boxDownload all the Genomic features and their annotations in GFF format
- Options:
- Do not generate feature for ncRNA genes (CDS genes only): If this is NOT selected, all annotated features will be included in the GFF file (chromosomes, tRNAs, rRNAs, repeat regions, etc). If it IS selected, only features coding for protein coding sequence (CDS) will be created.
- Include feature annotations (descriptive text; Geneontology; etc): All annotations for a feature will be included in the GFF file.
- Ensure the GFF Name tag is unique for each feature: Each feature Name Tag will have the type appended to it along with a unique number incremented from one:
- Do not propagate duplicate annotations to children: If selected, an additional check will be performed for each annotation. If a parent feature already contains the annotation, it will NOT be included in the child feature.
- For GFF "ID" and "Parent" tags, use unique: Specified whether each ID tag is a unique number or human readable name.
- Number: The GFF Exporter will start with ID 1 and increment it for each feature.
- Name: The GFF Exporter will using one of the names stored in CoGe for the feature and postpend a suffix based on the feature type (e.g. gene, mRNA, CDS, tRNA, exon) and a unique number.
- Examples: ID=AT3G01015.gene1;Name=AT3G01015.gene1;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025
##sequence-region 3 1 23459830 3 CoGe chromosome 1 23459830 . + . ID=3.chromosome1;Name=3.chromosome1;Alias=3;coge_fid=78616734 3 CoGe gene 1653 4159 . - . ID=AT3G01015.gene1;Name=AT3G01015.gene1;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640506;encoded_feature=mRNA 3 CoGe mRNA 1653 4159 . - . Parent=AT3G01015.gene1;ID=AT3G01015.mRNA1;Name=AT3G01015.mRNA1;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640506 3 CoGe exon 1653 1936 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.exon1;Name=AT3G01015.exon1;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640503 3 CoGe exon 2048 2142 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.exon2;Name=AT3G01015.exon2;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640503 3 CoGe exon 2223 2282 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.exon3;Name=AT3G01015.exon3;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640503 3 CoGe exon 2428 2526 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.exon4;Name=AT3G01015.exon4;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640503 3 CoGe exon 2690 2809 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.exon5;Name=AT3G01015.exon5;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640503 3 CoGe exon 2885 2977 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.exon6;Name=AT3G01015.exon6;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640503 3 CoGe exon 3064 3109 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.exon7;Name=AT3G01015.exon7;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640503 3 CoGe exon 3203 4159 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.exon8;Name=AT3G01015.exon8;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640503 3 CoGe CDS 1798 1936 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.CDS1;Name=AT3G01015.CDS1;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640508 3 CoGe CDS 2048 2142 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.CDS2;Name=AT3G01015.CDS2;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640508 3 CoGe CDS 2223 2282 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.CDS3;Name=AT3G01015.CDS3;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640508 3 CoGe CDS 2428 2526 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.CDS4;Name=AT3G01015.CDS4;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640508 3 CoGe CDS 2690 2809 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.CDS5;Name=AT3G01015.CDS5;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640508 3 CoGe CDS 2885 2977 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.CDS6;Name=AT3G01015.CDS6;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640508 3 CoGe CDS 3064 3109 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.CDS7;Name=AT3G01015.CDS7;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640508 3 CoGe CDS 3203 4017 . - . Parent=AT3G01015.mRNA1;ID=AT3G01015.CDS8;Name=AT3G01015.CDS8;Alias=AT3G01015,AT3G01015.1,Locus.13646,Model.15025;coge_fid=78640508 3 CoGe gene 4342 4818 . + . ID=AT3G01010.gene1;Name=AT3G01010.gene1;Alias=AT3G01010,AT3G01010.1,Locus.13644,Model.15056;coge_fid=78625288;encoded_feature=mRNA 3 CoGe mRNA 4342 4818 . + . Parent=AT3G01010.gene1;ID=AT3G01010.mRNA1;Name=AT3G01010.mRNA1;Alias=AT3G01010,AT3G01010.1,Locus.13644,Model.15056;coge_fid=78625288 3 CoGe exon 4342 4818 . + . Parent=AT3G01010.mRNA1;ID=AT3G01010.exon1;Name=AT3G01010.exon1;Alias=AT3G01010,AT3G01010.1,Locus.13644,Model.15056;coge_fid=78625287 3 CoGe CDS 4342 4818 . + . Parent=AT3G01010.mRNA1;ID=AT3G01010.CDS1;Name=AT3G01010.CDS1;Alias=AT3G01010,AT3G01010.1,Locus.13644,Model.15056;coge_fid=78625289
- Note: Each GFF annotation line contains the CoGe Feature ID. E.g.: CoGe_fid="78903869";
- You can use the feature ID to construct links to CoGe to access information about that feature: http://genomevolution.org/CoGe/FeatView.pl?fid=78903869
- Note: Each GFF annotation line contains the CoGe Feature ID. E.g.: CoGe_fid="78903869";
Additional Genome Options
- Click for Features: Link to generate a summary table of all features in the genome that will be displayed below the "Chromosome information" in a feature list
- OrganismView Link: This URL will regenerate the page with the selected genome pre-loaded. Useful for saving the information for later or sending it to someone else
- Add to Genome List: If you click this button, the genome will be added to a list of genomes. A popup box will appear with your current list of genomes.
- Owner functions: This buttons will appear if you are the owner of the genome
- Make Genome Private: Remove the genome from public view. Only users in User groups which have access to the genome may see the genome.
- Make Genome Public: Makes the genome viewable by anyone.
- Edit Genome Info: allows an owner of the genome to change the name, description, version of the genome. May also add a message to be displayed when the genome is viewed as well as specify a link to additional information about the genome. When this button is pressed, a popup dialog box will appear to allow a user to modify this information.
- Note: the link must be in the format of 'http://something.com/whatever/etc'
Dataset Information
Chromosome Information
Genomic Data
GC content
- Total
- Non-coding
Feature Lists
Links
Genome Viewer
Get Sequence
Linking to OrganismView
It is relatively easy to link directly into OrganismView to search for an organism or retrieve a specific organism. Please see Linking to OrganismView for more information.