CoGepedia:Community Portal: Difference between revisions
Created page with ''''Astrids and eurosids share a paleohexaploidy'''' |
|||
| (15 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
''' | ==Using GEvo to determine that the astrids and eurosids share the paleohexaploidy== | ||
[[File:grape-tomato-synteny.png|thumb|500px|Syntenic comparison using GEvo between grape and tomato showing a near-perfect 1:1 mapping of annotated grape genes to an region of tomato that has not annotations]] | |||
The image above shows a syntenic alignment between genomic regions of grape and tomato. Nearly every gene in grape has a syntenic match to tomato (the tomato genomic region is not annotated). This near-perfect one-to-one mapping of gene between fairly distantly related plant genomes (one from each of two major groups of eudicots -- eurosids and astrids) is the expected pattern if neither genome has undergone one or more independent [[whole genome duplication]] ([[WGD]]) events. In plants, following WGD, each duplicated genomic region undergoes diploidization, which reduces the total gene content of the genome to one which is more similar to the pre-duplicated ancestor genome. This process of gene loss is known as fractionation and results in distributing ancestral gene content over all genomic duplicates. If such a process were at work, we would not expect to see the near-perfect one-to-one mapping of gene content. Results can be regenerated and analysis resumed using http://tinyurl.com/qjpcvv. | |||
==SynMap comparison of Arabidopsis thaliana and Arabidopsis lyrata showing how syntenic regions derived from different whole genome duplications are comprised of syntelogs with characteristic different synonymous mutation rates== | |||
[[File:Master_3068_8.CDS-CDS.blastn_geneorder_D20_g10_A5.w2000.ks.png|thumb|500px|Syntenic dotplot between two Arabidopsis specis ''thaliana'' and ''lyrata'' showing synteny derived from speciation and a shared whole genome duplication event.]] | |||
This syntenic dotplot is between two Arabidopsis species, ''A. lyrata'' on the x-axis and ''A. thaliana'' on the y-axis. Each putative homologous match between two genes results in a gray dot being drawn on the plot. The results are analyzed by DAGChainer to identify syntenic regions and syntelogous gene pairs are analyzed by CODEML to calculate their synonymous mutation rate. This dotplot shows extensive stretches of synteny between these two genomes in a two-to-two relationship. Each genomic region is syntenic to two region in the other genome. One that is orthologous and derived from the speciation of the lineages, and the other from a shared whole genome duplication event. Although it is fairly obvious based on the size and extent of synteny as to which syntenic region is derived from speciation and WGD, the coloration of the syntelogs by synonymous mutation rate helps differentiate the events. Since the WGD happened prior to the speciation event, it is expected that genes retained from the WGD have had more time to undergo mutation, and will thus have a higher apparent synonymous mutation rate. Also, since both of these events created contemporaneous copies of all genes and chromosomes at once, those genes will have similar apparent rates of synonymous mutations. | |||
[[File:Master_3068_8.CDS-CDS.blastn_geneorder_D20_g10_A5.w2000.ks.hist.png|thumb|300px|Distribution of synonymous mutation rates calculated for syntelogs between ''Arabidopsis thaliana'' and ''Arabidopsis lyrata''. The first hump in the distribution is for syntelogs derived from speciation, the second from their shared whole genome duplication (WGD) event, and the third may be from a more ancient WGD, noise, or other genome duplication event.]] | |||
This is a histogram of synonymous mutation rates for the syntenic dotplot above. The left most hump are genes derived from the speciation of these lineages, the middle hump is derived from the most recent shared WGD, and the left hump may represent a more ancient WGD (or noise, or some other genomic duplication event). While these lineages do share at least two more ancient WGD, the data presented in this synonymous rate distribution are inconclusive. However, other types of comparative genomic approaches may be employed. | |||
For more information, please [[SynMap#Example_Results | see this example using SynMap.]] | |||
Latest revision as of 08:06, 2 February 2010
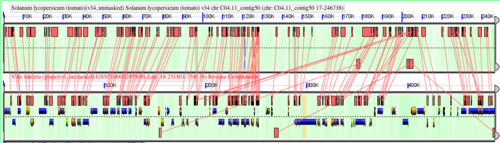
The image above shows a syntenic alignment between genomic regions of grape and tomato. Nearly every gene in grape has a syntenic match to tomato (the tomato genomic region is not annotated). This near-perfect one-to-one mapping of gene between fairly distantly related plant genomes (one from each of two major groups of eudicots -- eurosids and astrids) is the expected pattern if neither genome has undergone one or more independent whole genome duplication (WGD) events. In plants, following WGD, each duplicated genomic region undergoes diploidization, which reduces the total gene content of the genome to one which is more similar to the pre-duplicated ancestor genome. This process of gene loss is known as fractionation and results in distributing ancestral gene content over all genomic duplicates. If such a process were at work, we would not expect to see the near-perfect one-to-one mapping of gene content. Results can be regenerated and analysis resumed using http://tinyurl.com/qjpcvv.
SynMap comparison of Arabidopsis thaliana and Arabidopsis lyrata showing how syntenic regions derived from different whole genome duplications are comprised of syntelogs with characteristic different synonymous mutation rates

This syntenic dotplot is between two Arabidopsis species, A. lyrata on the x-axis and A. thaliana on the y-axis. Each putative homologous match between two genes results in a gray dot being drawn on the plot. The results are analyzed by DAGChainer to identify syntenic regions and syntelogous gene pairs are analyzed by CODEML to calculate their synonymous mutation rate. This dotplot shows extensive stretches of synteny between these two genomes in a two-to-two relationship. Each genomic region is syntenic to two region in the other genome. One that is orthologous and derived from the speciation of the lineages, and the other from a shared whole genome duplication event. Although it is fairly obvious based on the size and extent of synteny as to which syntenic region is derived from speciation and WGD, the coloration of the syntelogs by synonymous mutation rate helps differentiate the events. Since the WGD happened prior to the speciation event, it is expected that genes retained from the WGD have had more time to undergo mutation, and will thus have a higher apparent synonymous mutation rate. Also, since both of these events created contemporaneous copies of all genes and chromosomes at once, those genes will have similar apparent rates of synonymous mutations.

This is a histogram of synonymous mutation rates for the syntenic dotplot above. The left most hump are genes derived from the speciation of these lineages, the middle hump is derived from the most recent shared WGD, and the left hump may represent a more ancient WGD (or noise, or some other genomic duplication event). While these lineages do share at least two more ancient WGD, the data presented in this synonymous rate distribution are inconclusive. However, other types of comparative genomic approaches may be employed.
For more information, please see this example using SynMap.