CodeOn: Difference between revisions
Jump to navigation
Jump to search
Created page with '[[Image:CDSEvo-MG1655.png|thumb|right|600px|Screenshot of CDSEvo showing amino acid usage for all CDSs in Escherichia coli K12 MG1655. CDSs are binned according to their overall ...' |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
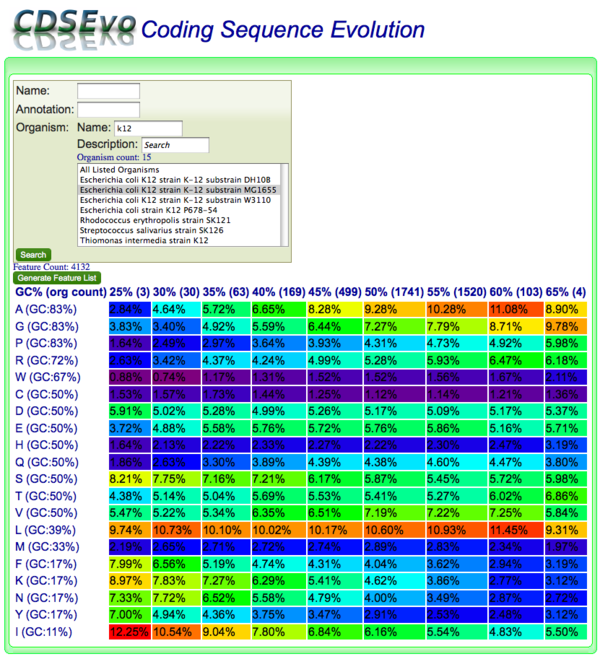
[[Image:CDSEvo-MG1655.png|thumb|right|600px|Screenshot of | [[Image:CDSEvo-MG1655.png|thumb|right|600px|Screenshot of CodeOn showing amino acid usage for all CDSs in Escherichia coli K12 MG1655. CDSs are binned according to their overall DNA GC content.]] | ||
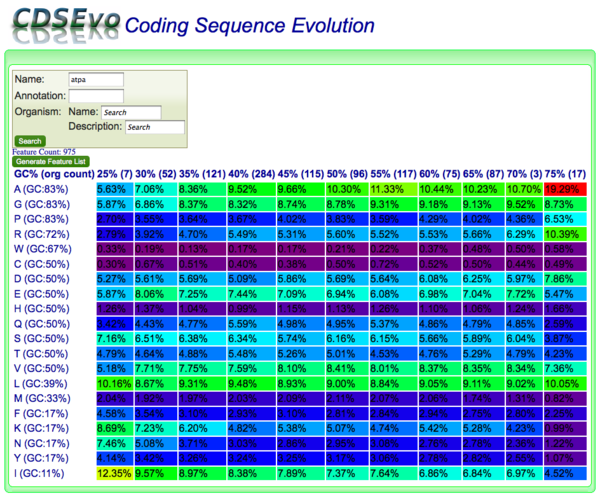
[[Image:CDSEvo-atpa.png|thumb|right|600px|Screenshot of | [[Image:CDSEvo-atpa.png|thumb|right|600px|Screenshot of CodeOn showing amino acid usage for all CDSs with "ATPA" in their name across all genomes in CoGe. ATPA is one of the highly conserved subunits of ATP synthase. CDSs are binned according to their overall DNA GC content.]] | ||
=Coding Sequence Evolution (CodeOn) | =Coding Sequence Evolution (CodeOn)= | ||
===Overview=== | ===Overview=== | ||
| Line 30: | Line 30: | ||
**Order: purple < blue < green < yellow < orange < red | **Order: purple < blue < green < yellow < orange < red | ||
===Linking to | ===Linking to CodeOn=== | ||
*fid= CoGe database feature id. Can have multiple specified either by fid=1&fid=2&fid=3 OR fid=1::2::3 | *fid= CoGe database feature id. Can have multiple specified either by fid=1&fid=2&fid=3 OR fid=1::2::3 | ||
Latest revision as of 18:39, 11 April 2010
Coding Sequence Evolution (CodeOn)
Overview
CodeOn allows you to search for CDS genomic features by name, annotation, and/or organism and display an amino acid usage table binned by the overall GC content of each CDS.
Usage
- Type in a name of a genomic feature
- Type in an annotation of a genomic feature
- Type in the name or description of an organism
Then press "search"
- When you search by genomic feature name and annotation, you can limit your search to a specific organism or set of organisms.
- If no genomic feature name or annotation is specified, but an organism or a group of organisms is, the search will get all CDSs from those organisms.
Results
The results table is organized such that:
- x-axis: CDSs are binned according to their GC content along
- y-axis: amino acids are listed according the GC content of their codons
Each cell contains:
- the relative percent usage of that amino acid for that bin
- is color-coded according to percent usage compared to all cells in the table:
- purple: lowest
- red: highest
- Order: purple < blue < green < yellow < orange < red
Linking to CodeOn
- fid= CoGe database feature id. Can have multiple specified either by fid=1&fid=2&fid=3 OR fid=1::2::3
- oid= CoGe database organism id.
- accn= Feature name
- anno= Feature annotation