Classical Maize Genes Examples: Difference between revisions
No edit summary |
|||
| (9 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
Interesting examples taken from the [[Classical Maize Genes|Classical Maize Genes]] dataset. | Interesting examples taken from the [[Classical Maize Genes|Classical Maize Genes]] dataset. | ||
== | The B73_refgen2 version of the classical maize gene list has been peer reviewed and is now published. Check it out! | ||
Schnable JC, Freeling M. "Genes Identified by Visible Mutant Phenotypes Show Increased Bias toward One of Two Subgenomes of Maize." PLoS One '''DOI:'''[http://dx.doi.org/10.1371/journal.pone.0017855 10.1371/journal.pone.0017855] | |||
A modified version of C1/Pl1 example shown below was included as a supplemental figure in the publication. | |||
__TOC__ | |||
== Local duplications of phytochrome A == | |||
The phytochromes are a class of genes that encode photoreceptors, proteins that help a plant tell the kind of light what light falls upon it. Since plants can't run away they need to adapt to their environment, and to adapt to their environment they must be able to perceive it. Most grasses are considered to have 3 phytochromes: A, B, and C. Maize still contains two copies of each gene from the maize whole genome duplication (A1, A2, B1, B2, C1, and C2). | The phytochromes are a class of genes that encode photoreceptors, proteins that help a plant tell the kind of light what light falls upon it. Since plants can't run away they need to adapt to their environment, and to adapt to their environment they must be able to perceive it. Most grasses are considered to have 3 phytochromes: A, B, and C. Maize still contains two copies of each gene from the maize whole genome duplication (A1, A2, B1, B2, C1, and C2). | ||
Looking at [http://tinyurl.com/yj3g4wz the syntenic panel for phytochrome A1 from maize], we can see that both Brachypodium and Sorghum contain a local duplicate of phytochrome A. Further examination shows that the two copies in sorghum (Sb01g009920 and Sb01g009930) are virtually identical, while one of the copies in Brachypodium (marked as PhyA?) has a 3' truncated three prime end. <br> | Looking at [http://tinyurl.com/yj3g4wz the syntenic panel for phytochrome A1 from maize], we can see that both Brachypodium and Sorghum contain a local duplicate of phytochrome A. Further examination shows that the two copies in sorghum (Sb01g009920 and Sb01g009930) are virtually identical as can be seen in [http://tinyurl.com/yghj3qh this comparison] to a GenBank Sorghum Phytochrome A sequence, while one of the copies in Brachypodium (marked as PhyA?) has a 3' truncated three prime end. <br> | ||
[[Image:Phyprime.png]] | [[Image:Phyprime.png]] | ||
== Pericarp color1 and Pericarp color2, local duplicate genes == | == Pericarp color1 and Pericarp color2, local duplicate genes == | ||
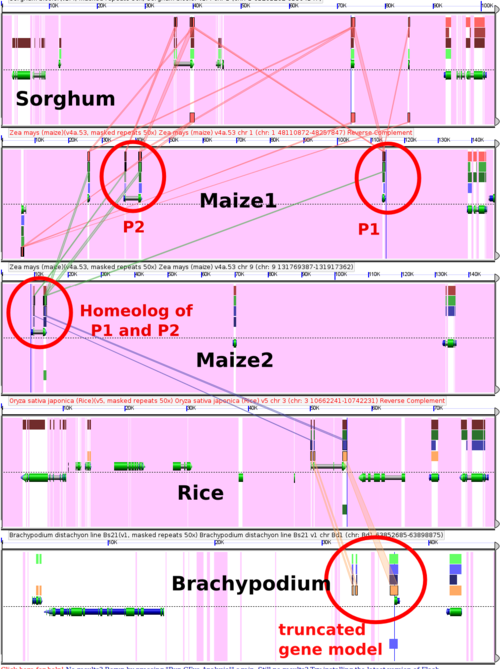
The pericarp color1 mutant dates back to at least the time of R. A. Emerson, the pericarp color2 mutant was discovered [http://www.plantcell.org/cgi/content/abstract/12/12/2311 more recently] and mapped to a nearby location in the maize genome. Here we can see that they are both members of an array of locally duplicated genes (or gene fragments) that is also present in Sorghum. Note that there is a single pericarp color-like gene in the homeologous region of the maize genome (created by the maize whole genome duplication). And see that the gene model in Brachypodium for the ortholog of P1 and P2 is truncated, but, once I turn off the default masking of non-CDS sequences, we can see that the rest of the gene is still there, just not annotated. | |||
The GEvo panel shown here can be regenerated at: http://tinyurl.com/ycp5hsb | |||
[[Image:Pericarps.png|500px]] | |||
== Colored aleurone1 and Purple plant1 created by the maize whole genome duplication == | |||
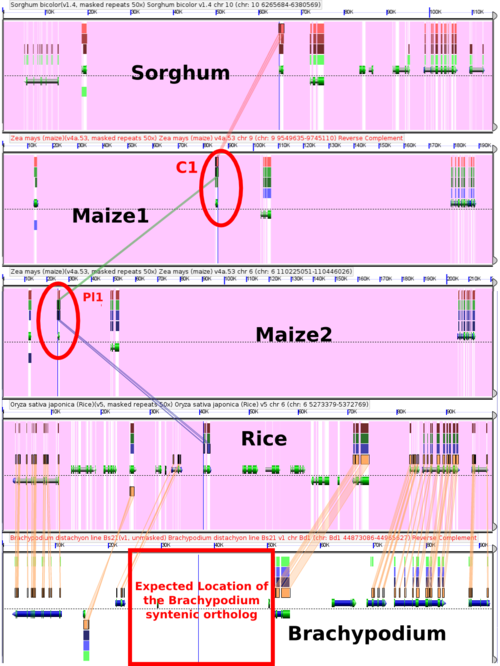
Colored aleurone1 and Purple plant1 are both genes with long histories in maize research and are involved in the regulation of anthocyanin biosynthesis. The two genes are also duplicates (homeologs) resulting from the maize whole genome duplication, with C1 located on Maize1 and Pl1 located on Maize2. From the included panel you can also see that region of brachypodium where we would expect to find an orthologous gene, based on synteny to rice, is empty. | |||
Note that I've removed all masking from the Brachypodium sequence (masked sequences in GEvo are shown as purple. | |||
The GEvo panel shown here can be regenerated at: http://tinyurl.com/yddlwor | |||
[[Image:C1pl1.png|500px]] | |||
Latest revision as of 19:40, 21 March 2011
Interesting examples taken from the Classical Maize Genes dataset.
The B73_refgen2 version of the classical maize gene list has been peer reviewed and is now published. Check it out!
Schnable JC, Freeling M. "Genes Identified by Visible Mutant Phenotypes Show Increased Bias toward One of Two Subgenomes of Maize." PLoS One DOI:10.1371/journal.pone.0017855
A modified version of C1/Pl1 example shown below was included as a supplemental figure in the publication.
Local duplications of phytochrome A
The phytochromes are a class of genes that encode photoreceptors, proteins that help a plant tell the kind of light what light falls upon it. Since plants can't run away they need to adapt to their environment, and to adapt to their environment they must be able to perceive it. Most grasses are considered to have 3 phytochromes: A, B, and C. Maize still contains two copies of each gene from the maize whole genome duplication (A1, A2, B1, B2, C1, and C2).
Looking at the syntenic panel for phytochrome A1 from maize, we can see that both Brachypodium and Sorghum contain a local duplicate of phytochrome A. Further examination shows that the two copies in sorghum (Sb01g009920 and Sb01g009930) are virtually identical as can be seen in this comparison to a GenBank Sorghum Phytochrome A sequence, while one of the copies in Brachypodium (marked as PhyA?) has a 3' truncated three prime end.
Pericarp color1 and Pericarp color2, local duplicate genes
The pericarp color1 mutant dates back to at least the time of R. A. Emerson, the pericarp color2 mutant was discovered more recently and mapped to a nearby location in the maize genome. Here we can see that they are both members of an array of locally duplicated genes (or gene fragments) that is also present in Sorghum. Note that there is a single pericarp color-like gene in the homeologous region of the maize genome (created by the maize whole genome duplication). And see that the gene model in Brachypodium for the ortholog of P1 and P2 is truncated, but, once I turn off the default masking of non-CDS sequences, we can see that the rest of the gene is still there, just not annotated.
The GEvo panel shown here can be regenerated at: http://tinyurl.com/ycp5hsb
Colored aleurone1 and Purple plant1 created by the maize whole genome duplication
Colored aleurone1 and Purple plant1 are both genes with long histories in maize research and are involved in the regulation of anthocyanin biosynthesis. The two genes are also duplicates (homeologs) resulting from the maize whole genome duplication, with C1 located on Maize1 and Pl1 located on Maize2. From the included panel you can also see that region of brachypodium where we would expect to find an orthologous gene, based on synteny to rice, is empty.
Note that I've removed all masking from the Brachypodium sequence (masked sequences in GEvo are shown as purple.
The GEvo panel shown here can be regenerated at: http://tinyurl.com/yddlwor