Ancestral Reconstruction Pipeline: Difference between revisions
No edit summary |
|||
| (47 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[Image:20140424 165903.jpg|thumb|right|200px|Pipeline]] [[Image:Photo.JPG|thumb|right|200px|Refactored pipeline]] | |||
*[[Images from hackathon]] | |||
*[[Notes on pipeline details]] | |||
=== Plan for refactoring === | |||
==== GetGenomes: remove config file, add option to specify output dir for output files ==== | |||
This program gets data ready for downstream processing | |||
* -d <directory of input synmap files> | |||
* -g gid1,gid2,gid3,gid4... <list of common separated coge genome ids> | |||
* -p p1,p2,p3,p4 <list of comma separated ploidy levels for genomes -- note these are paired ordered data with the -g option?> | |||
* -s < subgenome_file> | |||
* -o <output directory for SubGenomeInGeneOrder OrthologSets GenomeInString files> | |||
====GetContigInput==== | |||
This program gets data ready for ancestral ordering of genes by MWM | |||
*Remove config file dependency | |||
* -g gid1,gid2,gid3,gid4... <list of common separated coge genome ids> | |||
* -w w1,w2,w3,w4 <list of comma separated weights for genomes -- note these are paired ordered data with the -g option?> | |||
* -wa <threshold minimum adjacency score for keeping a contig. Called 'weightOfAdjacent' in original config file> | |||
* -i <input file: GenomesInString from program GetGenomes> | |||
* -o <output file name> | |||
Note: Output is now a tab delimited file with each line containing: vertex vertex weight . These data will be used by the MEMPython data below | |||
====MWMPython: http://jorisvr.nl/maximummatching.html needs command line options for ==== | |||
This program is a general tool for Maximum Weight Matching. First run is for ancestral gene ordering. Second run is for ancestral contig ordering | |||
* -i <input file or directory> | |||
** File type is a set of vertex vertex weight | |||
** note: if directory, will batch process all files | |||
* -o <outfile or directory> If no option is specified, the results go to STDOUT | |||
====GetContigOutputAndScaffoldInput==== | |||
This program maps ancestral contigs back to various genomes, gets their positions, and gets data formatted for a second MWM to generate ancestral ordered contigs | |||
Note: goal is to get everything onto the command line. Currently, several of the files are hardcoded | |||
*-mml <threshold minimum mapping length. The minimum number of "genes" mapping to a subgenome. Called 'minimumGeneGroupLength' in original config file> | |||
'''Note''': genomeInContigIndex is specified in the config file. This is the weighting for each subgenome. These values will be removed and the weights for genomes used instead: | |||
* -g gid1,gid2,gid3,gid4... <list of common separated coge genome ids> | |||
* -w w1,w2,w3,w4 <list of comma separated weights for genomes -- note these are paired ordered data with the -g option> | |||
*-co <configoutput file generated by the MWMPython program> | |||
*-s <subGenomesInGeneOrder file> | |||
*-gf <genomeInString file> | |||
* -o <output directory name> | |||
'''Note:''' Multiple output files, one for each ancestral chromosome bin | |||
'''Note:''' There are three sets of data that are needed for the subsequent steps: | |||
* Directory named "scaffolds". These tab delimited files with each line containing: vertex vertex weight. These data will be used by the MEMPython program | |||
* Directory named "binfiles". These files are used in the final program ScaffoldOutput to assign the correct contigs to the bins | |||
* File named "contig2genes.txt" This file is used to convert the contigs back to genes. These are 'syntenic gene sets'. | |||
====MWMPython==== | |||
Reuse of the same program listed above for ancestral ordering of contigs | |||
====ScaffoldOutput==== | |||
Putting all the output data back together for the final ancestral genome | |||
*-im <directory of MWM files> | |||
*-ib <directory composition of bin files> | |||
** This directory is called 'binfiles' when generated by GetContigOutputAndScaffoldInput | |||
*-cg <file containing the looking up of gene families comprising contigs | |||
**This file is named 'contig2genes.txt' when generated by GetContigOutputAndScaffoldInput | |||
*-o <output file of reconstructed genome> | |||
< | |||
Latest revision as of 01:46, 26 April 2014

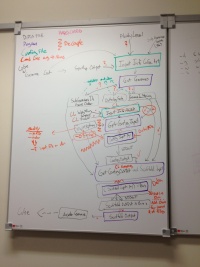
Plan for refactoring
GetGenomes: remove config file, add option to specify output dir for output files
This program gets data ready for downstream processing
- -d <directory of input synmap files>
- -g gid1,gid2,gid3,gid4... <list of common separated coge genome ids>
- -p p1,p2,p3,p4 <list of comma separated ploidy levels for genomes -- note these are paired ordered data with the -g option?>
- -s < subgenome_file>
- -o <output directory for SubGenomeInGeneOrder OrthologSets GenomeInString files>
GetContigInput
This program gets data ready for ancestral ordering of genes by MWM
- Remove config file dependency
- -g gid1,gid2,gid3,gid4... <list of common separated coge genome ids>
- -w w1,w2,w3,w4 <list of comma separated weights for genomes -- note these are paired ordered data with the -g option?>
- -wa <threshold minimum adjacency score for keeping a contig. Called 'weightOfAdjacent' in original config file>
- -i <input file: GenomesInString from program GetGenomes>
- -o <output file name>
Note: Output is now a tab delimited file with each line containing: vertex vertex weight . These data will be used by the MEMPython data below
MWMPython: http://jorisvr.nl/maximummatching.html needs command line options for
This program is a general tool for Maximum Weight Matching. First run is for ancestral gene ordering. Second run is for ancestral contig ordering
- -i <input file or directory>
- File type is a set of vertex vertex weight
- note: if directory, will batch process all files
- -o <outfile or directory> If no option is specified, the results go to STDOUT
GetContigOutputAndScaffoldInput
This program maps ancestral contigs back to various genomes, gets their positions, and gets data formatted for a second MWM to generate ancestral ordered contigs Note: goal is to get everything onto the command line. Currently, several of the files are hardcoded
- -mml <threshold minimum mapping length. The minimum number of "genes" mapping to a subgenome. Called 'minimumGeneGroupLength' in original config file>
Note: genomeInContigIndex is specified in the config file. This is the weighting for each subgenome. These values will be removed and the weights for genomes used instead:
- -g gid1,gid2,gid3,gid4... <list of common separated coge genome ids>
- -w w1,w2,w3,w4 <list of comma separated weights for genomes -- note these are paired ordered data with the -g option>
- -co <configoutput file generated by the MWMPython program>
- -s <subGenomesInGeneOrder file>
- -gf <genomeInString file>
- -o <output directory name>
Note: Multiple output files, one for each ancestral chromosome bin
Note: There are three sets of data that are needed for the subsequent steps:
- Directory named "scaffolds". These tab delimited files with each line containing: vertex vertex weight. These data will be used by the MEMPython program
- Directory named "binfiles". These files are used in the final program ScaffoldOutput to assign the correct contigs to the bins
- File named "contig2genes.txt" This file is used to convert the contigs back to genes. These are 'syntenic gene sets'.
MWMPython
Reuse of the same program listed above for ancestral ordering of contigs
ScaffoldOutput
Putting all the output data back together for the final ancestral genome
- -im <directory of MWM files>
- -ib <directory composition of bin files>
- This directory is called 'binfiles' when generated by GetContigOutputAndScaffoldInput
- -cg <file containing the looking up of gene families comprising contigs
- This file is named 'contig2genes.txt' when generated by GetContigOutputAndScaffoldInput
- -o <output file of reconstructed genome>