Creosote: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| (18 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Twig2Genome Notes== | |||
[[Twig2Genome]] | |||
==Assembly== | |||
[[Creosote Assembly]] | |||
==Loading into CoGe== | |||
SoapDeNovo assemly: 1,570,116 contigs (370MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12183 ('''Note:''' too many contigs to process and visualize in SynMap) | |||
SoapDeNovo assembly of contigs >= 2000nt: 6,976 contigs (20MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12185 | |||
ABySS assembly (bpsize=64, 2kb minimum contig size): 122,972 contigs (392MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12275 | |||
* | *Syntenic Path Assembly to peach: http://genomevolution.org/r/3xmq | ||
Velvet assembly 515,190 contigs (241MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12245 | |||
* | |||
CLC4 assembly 685,475 contigs (508MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12244 | |||
*Syntenic Path Assembly to peach: http://genomevolution.org/r/3xpd | |||
==[[Syntenic Path Assembly]]== | |||
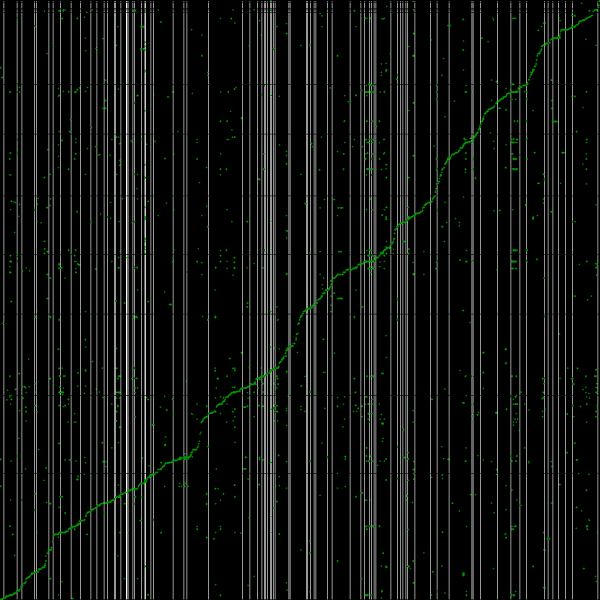
[[File:Master 12185 8400.genomic-CDS.lastz.dag.go c20 D20 g10 A2.aligncoords.gcoords ct0.w1000.ass2.cs1.csoS.nsd.png|thumb|600px|left|Syntenic path assembly with SynMap of creosote (x-axis) and peach (y-axis). Results may be regenerated at: http://genomevolution.org/r/3w95]] | |||
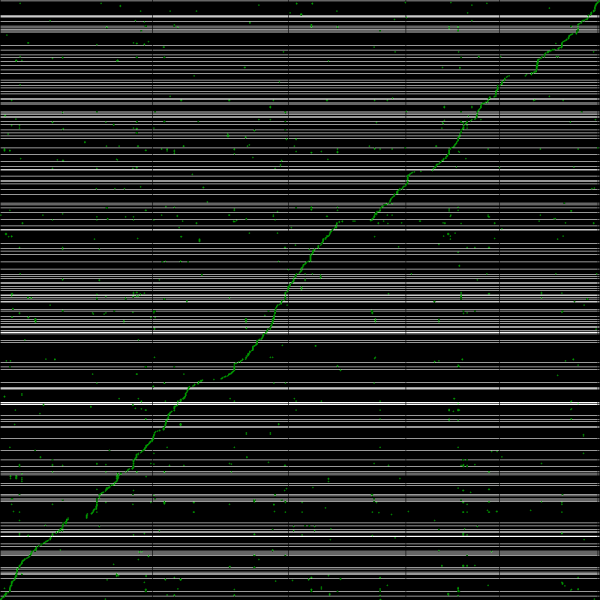
[[File:Master 11022 12185.CDS-genomic.lastz.dag.go c20 D20 g10 A2.aligncoords.gcoords ct0.w1000.ass2.cs1.csoS.nsd.png|thumb|600px|left|Syntenic path assembly with SynMap of creosote (y-axis) and Arabidopsis thaliana Col-0 (x-axis). Results may be regenerated at: http://genomevolution.org/r/3w96]] | |||
==Pseudo-Assembly== | |||
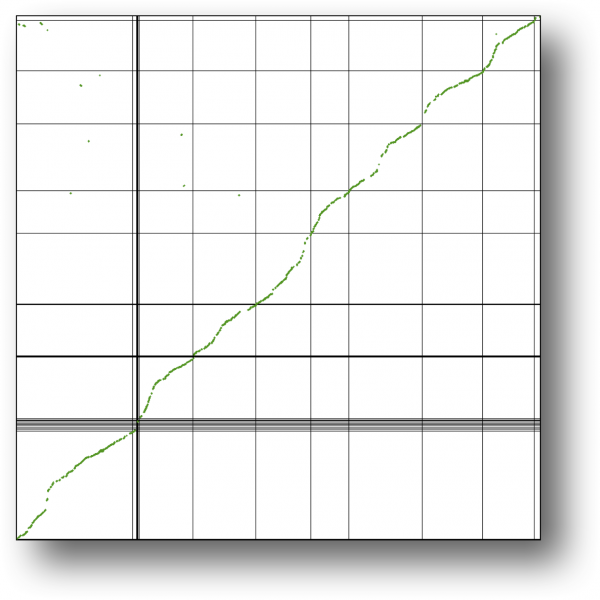
[[File:Pseudo-assembly-creosote.png|thumb|600px|left|Pseudo-assembly of creosote (x-axis) using the peach (y-axis) genome. Syntenic comparison to the peach genome.]] | |||
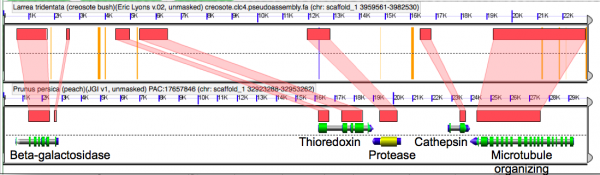
[[File:Screen Shot 2014-05-14 at 8.33.27 AM.png|thumb|600px|left|Microsynteny analysis of the pseudo-assembled creosote genome to the peach genome. Orange bars are unsequenced Ns that represent contigs glued together in creosote by the [[syntenic path assembly]] method. Note the concordance of gene model coding sequences.]] | |||
Latest revision as of 13:35, 14 May 2014
Twig2Genome Notes
Assembly
Loading into CoGe
SoapDeNovo assemly: 1,570,116 contigs (370MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12183 (Note: too many contigs to process and visualize in SynMap)
SoapDeNovo assembly of contigs >= 2000nt: 6,976 contigs (20MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12185
ABySS assembly (bpsize=64, 2kb minimum contig size): 122,972 contigs (392MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12275
- Syntenic Path Assembly to peach: http://genomevolution.org/r/3xmq
Velvet assembly 515,190 contigs (241MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12245
CLC4 assembly 685,475 contigs (508MB): http://genomevolution.org/CoGe/OrganismView.pl?dsgid=12244
- Syntenic Path Assembly to peach: http://genomevolution.org/r/3xpd
Syntenic Path Assembly
Pseudo-Assembly