Identifying SNPs: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
mNo edit summary |
||
| (10 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
CoGe has a built-in SNP finding pipeline! To find SNPs in an existing alignment-type experiment, select the "Find SNPs" analysis tool in the [ExperimentView] page. | CoGe has a built-in SNP finding pipeline! To find SNPs in an existing alignment-type experiment, select the "Find SNPs" analysis tool in the [[ExperimentView]] page. | ||
[File:ExperimentView_findSNPs.png|thumb|right|500px| | [[File:ExperimentView_findSNPs.png|thumb|right|500px|Find SNPs function (highlighted in red)]] | ||
An alignment-type experiment in CoGe is created by loading a BAM file. See [[LoadExperiment]] for instruction on how to do this. | |||
The CoGe SNP finding pipeline detects SNPs that meet the following criteria (these values are configurable): | |||
* minimum read depth of 10 (of any base quality value) | |||
* minimum high-quality allele count of 4 (high quality is PHRED >= 20) | |||
* minimum allele frequency of 10%. | |||
The SAMtools mpileup function is used to generate read depth counts for the BAM file. | |||
We are working to incorporate other SNP-finding programs into CoGe. Let us know if you'd like your favorite tool added! | |||
[[File:SNP_track.png|thumb|right|300px|Resulting SNP track (highlighted in red)]] | |||
Latest revision as of 16:52, 29 February 2016
CoGe has a built-in SNP finding pipeline! To find SNPs in an existing alignment-type experiment, select the "Find SNPs" analysis tool in the ExperimentView page.
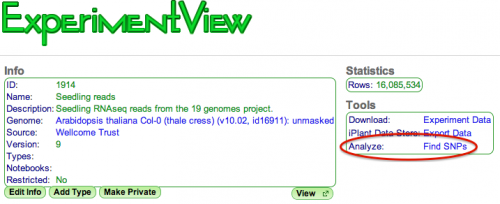
An alignment-type experiment in CoGe is created by loading a BAM file. See LoadExperiment for instruction on how to do this.
The CoGe SNP finding pipeline detects SNPs that meet the following criteria (these values are configurable):
- minimum read depth of 10 (of any base quality value)
- minimum high-quality allele count of 4 (high quality is PHRED >= 20)
- minimum allele frequency of 10%.
The SAMtools mpileup function is used to generate read depth counts for the BAM file.
We are working to incorporate other SNP-finding programs into CoGe. Let us know if you'd like your favorite tool added!
