Plasmodia comparative genomics: Difference between revisions
No edit summary |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[Image:Master_7949_2465.CDS-CDS.blastn.dag.go_c4_D20_g10_A5.aligncoords.gcoords_ct0.w1200.png|thumb|right|600px|Syntenic dotplot of Plasmodium falciparum (x-axis) and Plasmodium knowlesi (y-axis). Results can be regenerated at: http://genomevolution.org/CoGe/SynMap.pl?dsgid1=7949;dsgid2=2465;c=4;D=20;g=10;A=5;Dm=0;gm=0;w=0;b=1;ft1=1;ft2=1;do1=1;do2=1;do=40;dt=geneorder]] | [[Image:Master_7949_2465.CDS-CDS.blastn.dag.go_c4_D20_g10_A5.aligncoords.gcoords_ct0.w1200.png|thumb|right|600px|Syntenic dotplot of Plasmodium falciparum (x-axis) and Plasmodium knowlesi (y-axis). Results can be regenerated at: http://genomevolution.org/CoGe/SynMap.pl?dsgid1=7949;dsgid2=2465;c=4;D=20;g=10;A=5;Dm=0;gm=0;w=0;b=1;ft1=1;ft2=1;do1=1;do2=1;do=40;dt=geneorder]] | ||
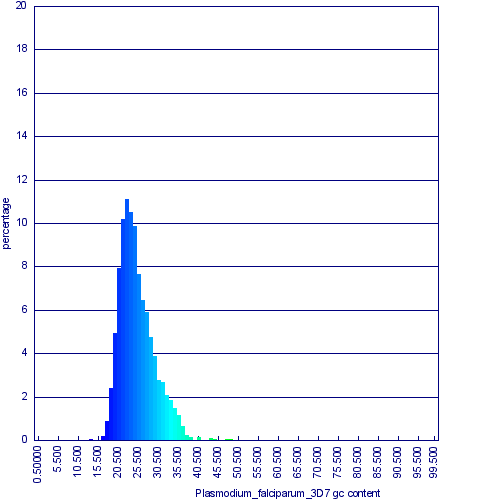
[[Image:Plasmodium falciparum cds gc content histogram.png|thumb|right|600px|Histogram of CDS GC content for Plasmodium falciparum. Generated by [[OrganismView]] ]] | [[Image:Plasmodium falciparum cds gc content histogram.png|thumb|right|600px|Histogram of CDS GC content for ''Plasmodium falciparum''. Generated by [[OrganismView]] ]] | ||
[[Image:Plasmodium knowlesi cds gc content histogram.png|thumb|right|600px|Histogram of CDS GC content for Plasmodium knowlesi. Generated by [[OrganismView]] ]] | [[Image:Plasmodium knowlesi cds gc content histogram.png|thumb|right|600px|Histogram of CDS GC content for ''Plasmodium knowlesi''. Generated by [[OrganismView]] ]] | ||
[[Image:Plasmodium-codon-substitution-matirx.png|thumb|right|600px|Log-odds score substitution matrix of codons between Plasmodium falciparum (x-axis) and Plasmodium knowlesi (y-axis). P. falciparum is a low-GC genome and P. | [[Image:Plasmodium-codon-substitution-matirx.png|thumb|right|600px|Log-odds score substitution matrix of codons between ''Plasmodium falciparum'' (x-axis) and ''Plasmodium knowlesi'' (y-axis). ''P. falciparum'' is a low-GC genome and ''P. knowlesi'' is a mid-GC genome.]] | ||
==Abstract== | =='''''Abstract'''''== | ||
''Plasmodium'' parasites have unique genomic features that make them an interesting case study in comparative genomics. In the last decade, the number of genome sequences for species of the ''Plasmodium'' genus has markedly increased. The availability of multiple ''Plasmodium'' genomes open the possibility to explore the genus genomic features and characteristics, and how these are shaped by evolutionary relationships. As thus, open-ended comparative analysis workflows represent a relevant approach to the study of parasites of the genus ''Plasmodium''. Many of GoGe‘s tools and services can be used to perform individual comparative analysis or in combination to evaluate evolutionary hypothesis. In the following pages, we will highlight the use of these tools on the case study of ''Plasmodium'' spp. | |||
*<span style="color:#006F00">'''Remain on this page for a quick overview of ''Plasmodium'' comparative genomics using CoGe.'''</span> | |||
*<span style="color:#006F00">'''For in-depth analyses workflows of ''Plasmodium'' genomes follow these links:'''</span> | |||
::*<span style="color:#006F00">Main page -</span> [[Using_CoGe_for_the_analysis_of_Plasmodium_spp]] | |||
::*<span style="color:#006F00">Workflow 1 -</span> [[Plasmodium analysis workflow 1: Tools that evaluate genomic properties and amino acid usage]] | |||
::*<span style="color:#006F00">Workflow 2 -</span> [[Plasmodium analysis workflow 2: Tools for the syntenic analysis of whole genomes and microsyntenic regions]] | |||
::*<span style="color:#006F00">Workflow 3 -</span> [[Plasmodium analysis workflow 3: Tools useful on the study of multigene families]] | |||
==Analysis== | =='''''Analysis'''''== | ||
The genomes of two Plasmodium species, falciparum and knowlesi are structurally very similar to one another: | The genomes of two Plasmodium species, falciparum and knowlesi are structurally very similar to one another: | ||
| Line 17: | Line 29: | ||
|Organism || Chromosome count || Genome Length || CDS count || Genome GC content || CDS GC content || CDS Wobble position content || non-coding GC content | |Organism || Chromosome count || Genome Length || CDS count || Genome GC content || CDS GC content || CDS Wobble position content || non-coding GC content | ||
|- | |- | ||
|Plasmodium falicparum || 14 || 22,860,235 bp || 5267 || 19.88% || 23.72% || 17.30% || 14.58% | |''Plasmodium falicparum'' || 14 || 22,860,235 bp || 5267 || 19.88% || 23.72% || 17.30% || 14.58% | ||
|- | |- | ||
|Plasmodium knowlesi || 14 || 23,462,187 bp || 5102 || 38.94% || 40.23% || 45.56% || 35.12% | |''Plasmodium knowlesi'' || 14 || 23,462,187 bp || 5102 || 38.94% || 40.23% || 45.56% || 35.12% | ||
|} | |} | ||
Their physical structure is also very similar, as can be seen in a syntenic dotplot of their genomes. However, their GC content is very different. ''P. falciparum''‘s overall GC content is 23% while ''P. knowlesi'' is 39%. Based on the similarities of their genomes' structures, this change in GC content is relatively recent, occurring after their lineages diverged between 2,000,000-10,000 years ago <ref>[http://www.pnas.org/content/early/2009/07/31/0907740106.abstract Rich et al. The origin of malignant malaria (2009) PNAS]</ref>. This change in their overall GC content is reflected in histograms of their respective [[CDS]] sequences, and their underlying codon and amino acid usages. Using syntenic gene pairs identified by their whole genome syntenic dotplot, protein alignments were generated and back translated to codon sequence alignments, and their entire data-set was used to calculate the log-odds score frequency of codon substitutions <ref>[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC50453/?tool=pubmed Henikoff and Henikoff. Amino acid substitution matrices from protein blocks. (1992) PNAS]</ref>. This substitution matrix is not symmetric. Each codon in each species has a different likelihood of being substituted than it being substituted back. This is a reflection of the apparent directionality in the GC content change. | |||
{{reflist}} | {{reflist}} | ||
Latest revision as of 20:48, 14 February 2017



Abstract
Plasmodium parasites have unique genomic features that make them an interesting case study in comparative genomics. In the last decade, the number of genome sequences for species of the Plasmodium genus has markedly increased. The availability of multiple Plasmodium genomes open the possibility to explore the genus genomic features and characteristics, and how these are shaped by evolutionary relationships. As thus, open-ended comparative analysis workflows represent a relevant approach to the study of parasites of the genus Plasmodium. Many of GoGe‘s tools and services can be used to perform individual comparative analysis or in combination to evaluate evolutionary hypothesis. In the following pages, we will highlight the use of these tools on the case study of Plasmodium spp.
- Remain on this page for a quick overview of Plasmodium comparative genomics using CoGe.
- For in-depth analyses workflows of Plasmodium genomes follow these links:
- Main page - Using_CoGe_for_the_analysis_of_Plasmodium_spp
Analysis
The genomes of two Plasmodium species, falciparum and knowlesi are structurally very similar to one another:
| Organism | Chromosome count | Genome Length | CDS count | Genome GC content | CDS GC content | CDS Wobble position content | non-coding GC content |
| Plasmodium falicparum | 14 | 22,860,235 bp | 5267 | 19.88% | 23.72% | 17.30% | 14.58% |
| Plasmodium knowlesi | 14 | 23,462,187 bp | 5102 | 38.94% | 40.23% | 45.56% | 35.12% |
Their physical structure is also very similar, as can be seen in a syntenic dotplot of their genomes. However, their GC content is very different. P. falciparum‘s overall GC content is 23% while P. knowlesi is 39%. Based on the similarities of their genomes' structures, this change in GC content is relatively recent, occurring after their lineages diverged between 2,000,000-10,000 years ago [1]. This change in their overall GC content is reflected in histograms of their respective CDS sequences, and their underlying codon and amino acid usages. Using syntenic gene pairs identified by their whole genome syntenic dotplot, protein alignments were generated and back translated to codon sequence alignments, and their entire data-set was used to calculate the log-odds score frequency of codon substitutions [2]. This substitution matrix is not symmetric. Each codon in each species has a different likelihood of being substituted than it being substituted back. This is a reflection of the apparent directionality in the GC content change.