Collinear gene order: Difference between revisions
Jump to navigation
Jump to search
Created page with 'thumb|600px|right|Syntenic comparison of two regions from the genome of Arabidopsis thaliana using [[GEvo. This genome underwent a whole genome duplic...' |
No edit summary |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[Image:Gevo-synteny.png|thumb|600px|right|Syntenic comparison of two regions from the genome of Arabidopsis thaliana using [[GEvo]]. This genome underwent a whole genome duplication event which created a copy of every genomic region. Over evolutionary time, many of the duplicated genes were lost by a process known as [[fractionation]]. However, many duplicated genes have been retained in duplicate and their collinear arrangement in the genome is evidence for synteny. Results can be regenerated at: http://tinyurl.com/noul6b]] | [[Image:Gevo-synteny.png|thumb|600px|right|Syntenic comparison of two regions from the genome of Arabidopsis thaliana using [[GEvo]]. This genome underwent a whole genome duplication event which created a copy of every genomic region. Over evolutionary time, many of the duplicated genes were lost by a process known as [[fractionation]]. However, many duplicated genes have been retained in duplicate and their collinear arrangement in the genome is evidence for synteny. Results can be regenerated at: http://tinyurl.com/noul6b]] | ||
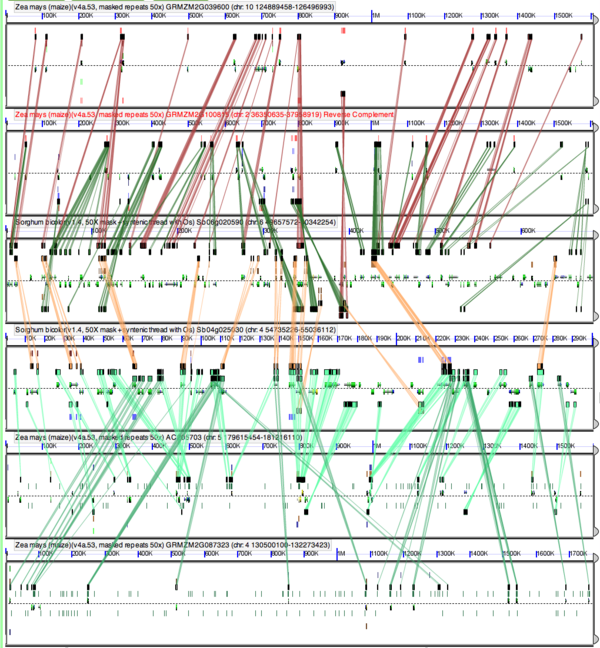
[[Image:Zm-Sb-6way.png|thumb|right|600px|High-resolution sequence comparison using [[GEvo]] of syntenic regions from maize and sorghum, including the pre-grass whole genome duplication event, the divergence of their lineages, and the maize-specific whole genome duplication event. These events create six syntenic regions identified each by a panel, 4 from maize (top two panels and bottom two panels) and 2 from sorghum (middle two panels). Results can be regenerated at: http://tinyurl.com/ycszpqs]] | |||
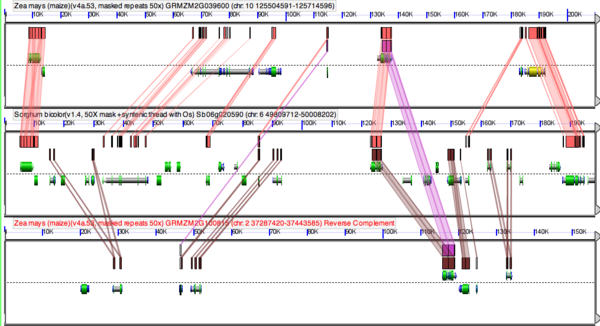
[[Image:Maize-sorghum-fractionation.png|thumb|right|600px|High-resolution sequence comparison using [[GEvo]] of a genomic region from sorghum and its two orthologous syntenic regions from maize. The sorghum region is the middle panel. Notice the strong pattern of collinear homologous genes between sorghum and both maize regions, while the maize regions only share two homologous genes with each other. This is due to the process of [[fractionation]] following the maize-specific whole genome duplication events. Results can be regenerated at: http://tinyurl.com/ydbbyeu]] | |||
[[Syntenic regions]], or genomic regions derived from a common ancestral genomic region, are inferred by identifying a collinear series of putatively homologous genes among two or more genomic regions. The inference is that the collinear order is the results of common ancestry rather than due to random chance. | [[Syntenic regions]], or genomic regions derived from a common ancestral genomic region, are inferred by identifying a collinear series of putatively homologous genes among two or more genomic regions. The inference is that the collinear order is the results of common ancestry rather than due to random chance. | ||
===Examples of collinear gene arrangement=== | |||
*[[GEvo-4at-cp-vv | Among the eurosids: Arabidopsis, papaya, and grapevine]] | |||
*[[Maize_Sorghum_Syntenic_dotplot | Maize and sorghum]] | |||
*[[Syntenic comparison of Arabidopsis thaliana and Arabidopsis lyrata | Arabidopsis thaliana and Arabidopsis lyrata]] | |||
*[[x-alignment| Various bacterial genomes]] | |||
Latest revision as of 01:47, 9 February 2010

Syntenic regions, or genomic regions derived from a common ancestral genomic region, are inferred by identifying a collinear series of putatively homologous genes among two or more genomic regions. The inference is that the collinear order is the results of common ancestry rather than due to random chance.