Cannabis sativa cultivar Chemdawg (marijuana): Difference between revisions
No edit summary |
|||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
This genome was sequenced by [http://www.medicinalgenomics.com/ Medicinal Genomics] (located in the Netherlands). It was sequenced with one lane of the Illumina HiSeq (2x100) platform and assembled with CLCbio’s workbench. Additional information about the assembly and genome may be found: http://www.medicinalgenomics.com/the-c-sativa-genome/ | This genome was sequenced by [http://www.medicinalgenomics.com/ Medicinal Genomics] (located in the Netherlands). It was sequenced with one lane of the Illumina HiSeq (2x100) platform and assembled with CLCbio’s workbench. Additional information about the assembly and genome may be found: http://www.medicinalgenomics.com/the-c-sativa-genome/ | ||
Cannabis is a member of the plant order Rosales. Of sequenced genomes in that order, the peach genome is a [[Why peach and grape genomes are peachy! | fantastic comparator]]. The reason for this is due to its high-quality sequence and assembly, | '''You can access Cannabis in CoGe:''' http://genomevolution.org/CoGe/OrganismView.pl?oid=33804 | ||
Cannabis is a member of the plant order Rosales. Of sequenced genomes in that order, the peach genome is a [[Why peach and grape genomes are peachy! | fantastic comparator]]. The reason for this is due to its high-quality sequence and assembly, and its genomic evolutionary history that does not contain any whole genome duplication event subsequent to the [[eudicot paleohexaploidy]] shared by nearly all dicots (at least the eurosids and the astrids). As such, its genome structure is probably very similar to the common ancestor of order Rosales, and perhaps the eudicots as a whole. This likely ancestral state of the peach genome makes it quite suitable for generating a [[pseudoassembly]] of highly fractured, low quality genome assemblies such as this Cannabis genome. CoGe's tool [[SynMap]] has an algorithm to tile contigs along any other "reference" genome in CoGe. | |||
The [[Syntenic path assembly]] of Cannabis to the peach genome may be viewed: http://genomevolution.org/wiki/index.php/Syntenic_path_assembly#Cannabis_sativa_.28marijuana.29_v._Prunus_persica_.28peach.29 | The [[Syntenic path assembly]] of Cannabis to the peach genome may be viewed: http://genomevolution.org/wiki/index.php/Syntenic_path_assembly#Cannabis_sativa_.28marijuana.29_v._Prunus_persica_.28peach.29 | ||
| Line 9: | Line 11: | ||
== [[Syntenic path assembly]] and syntenic dotplots of the Cannabis sativa (marijuana) v. Prunus persica (peach) == | == [[Syntenic path assembly]] and syntenic dotplots of the Cannabis sativa (marijuana) v. Prunus persica (peach) == | ||
[[SynMap]] has the ability to tile a contig level assembly against any other genome in CoGe using synteny. These [[pseudoassemblies]] can be printed out and then reloaded into CoGe. | [[SynMap]] has the ability to tile a contig level assembly against any other genome in CoGe using synteny. These [[pseudoassemblies]] can be printed out and then reloaded into CoGe. Regardless of how unreal and inaccurate pseudoassemblies are to the actual genome structure of an organism, having such high-order assemblies permits using CoGe's other tools to easily compare genomic regions. This method can rapidly turn a 170,000 piece genome into something useful for comparative genomics. | ||
<gallery widths =500px heights=500px compation="Syntenic Path Assembly in SynMap of two genomes in the order Rosales" perrow=2> | <gallery widths =500px heights=500px compation="Syntenic Path Assembly in SynMap of two genomes in the order Rosales" perrow=2> | ||
Latest revision as of 15:11, 24 August 2011
This genome was sequenced by Medicinal Genomics (located in the Netherlands). It was sequenced with one lane of the Illumina HiSeq (2x100) platform and assembled with CLCbio’s workbench. Additional information about the assembly and genome may be found: http://www.medicinalgenomics.com/the-c-sativa-genome/
You can access Cannabis in CoGe: http://genomevolution.org/CoGe/OrganismView.pl?oid=33804
Cannabis is a member of the plant order Rosales. Of sequenced genomes in that order, the peach genome is a fantastic comparator. The reason for this is due to its high-quality sequence and assembly, and its genomic evolutionary history that does not contain any whole genome duplication event subsequent to the eudicot paleohexaploidy shared by nearly all dicots (at least the eurosids and the astrids). As such, its genome structure is probably very similar to the common ancestor of order Rosales, and perhaps the eudicots as a whole. This likely ancestral state of the peach genome makes it quite suitable for generating a pseudoassembly of highly fractured, low quality genome assemblies such as this Cannabis genome. CoGe's tool SynMap has an algorithm to tile contigs along any other "reference" genome in CoGe.
The Syntenic path assembly of Cannabis to the peach genome may be viewed: http://genomevolution.org/wiki/index.php/Syntenic_path_assembly#Cannabis_sativa_.28marijuana.29_v._Prunus_persica_.28peach.29
This shows the Cannabis genome sequence contains nearly the entire gene content of Peach.
Syntenic path assembly and syntenic dotplots of the Cannabis sativa (marijuana) v. Prunus persica (peach)
SynMap has the ability to tile a contig level assembly against any other genome in CoGe using synteny. These pseudoassemblies can be printed out and then reloaded into CoGe. Regardless of how unreal and inaccurate pseudoassemblies are to the actual genome structure of an organism, having such high-order assemblies permits using CoGe's other tools to easily compare genomic regions. This method can rapidly turn a 170,000 piece genome into something useful for comparative genomics.
-
Syntenic dotplot by [SynMap]. X-axis Cannabis sativa (marijuana); Y-axis Prunus persica (peach). Results may be regenerated: http://genomevolution.org/r/3wz3
-
Syntenic dotplot by [SynMap]. X-axis Cannabis sativa (marijuana); Y-axis Prunus persica (peach). The Cannabis genome was pseudoassembled against the peach genome using SynMap's Syntenic path assembly algorithm. The assembled genome was reloaded into CoGe and compared against the peach genome. Results may be regenerated: http://genomevolution.org/r/3x1o
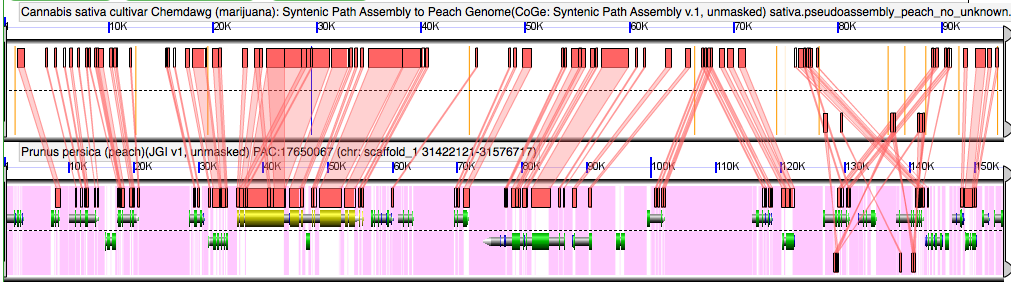
GEvo analysis of pseudoassembly of Cannabis genome to peach
Note that the pseudoassembled contigs in Cannabis are identifiable by orange bars in the background of the Cannabis panel. These orange bars are the 100 Ns used to join neighboring contigs. Results may be regenerated at http://genomevolution.org/r/3x1u .
![Syntenic dotplot by [SynMap]. X-axis Cannabis sativa (marijuana); Y-axis Prunus persica (peach). Results may be regenerated: http://genomevolution.org/r/3wz3](/wiki/images/thumb/f/f5/Master_12243_8400.genomic-CDS.lastz.dag.go_c4_D20_g10_A2.aligncoords.gcoords_ct0.w500.ass2.cs1.csoS.nsd.png/67px-Master_12243_8400.genomic-CDS.lastz.dag.go_c4_D20_g10_A2.aligncoords.gcoords_ct0.w500.ass2.cs1.csoS.nsd.png)
![Syntenic dotplot by [SynMap]. X-axis Cannabis sativa (marijuana); Y-axis Prunus persica (peach). The Cannabis genome was pseudoassembled against the peach genome using SynMap's Syntenic path assembly algorithm. The assembled genome was reloaded into CoGe and compared against the peach genome. Results may be regenerated: http://genomevolution.org/r/3x1o](/wiki/images/thumb/5/54/Master_12259_8400.genomic-CDS.lastz.dag.go_c4_D20_g10_A5.aligncoords.gcoords_ct0.w500.cs1.csoN.nsd.png/68px-Master_12259_8400.genomic-CDS.lastz.dag.go_c4_D20_g10_A5.aligncoords.gcoords_ct0.w500.cs1.csoN.nsd.png)