Utricularia and Genlisea pseudoassembly: Difference between revisions
Jump to navigation
Jump to search
Created page with 'thumb|center|600px|GEvo analysis of [[pseudoassembly Utricularia to tomato. The pseudoassembly of Utricularia is based on the [...' |
No edit summary |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Overview== | |||
[[Pseudoassemblies]] for both Utricularia and Genlisea are generated in [[SynMap]] using the [[Syntenic Path Assembly]] to the tomato genome. | |||
==High resolution analyses== | |||
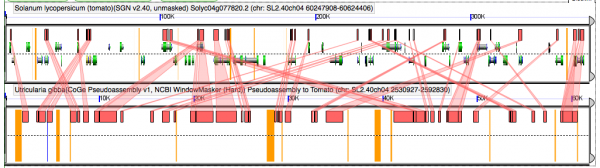
[[File:Screen Shot 2012-02-06 at 6.26.10 AM2.png|thumb|center|600px|GEvo analysis of [[pseudoassembly]] Utricularia to tomato. The pseudoassembly of Utricularia is based on the [[syntenic path assembly]] to tomato. As such, duplicated utricularia genomic regions will appear next to each other. If Utricularia has had an independent [[WGD]] followed by fractionation, these neighboring pieces will show [[intercalated synteny]] to tomato. Results may be regenerated at http://genomevolution.org/r/4h2c]] | [[File:Screen Shot 2012-02-06 at 6.26.10 AM2.png|thumb|center|600px|GEvo analysis of [[pseudoassembly]] Utricularia to tomato. The pseudoassembly of Utricularia is based on the [[syntenic path assembly]] to tomato. As such, duplicated utricularia genomic regions will appear next to each other. If Utricularia has had an independent [[WGD]] followed by fractionation, these neighboring pieces will show [[intercalated synteny]] to tomato. Results may be regenerated at http://genomevolution.org/r/4h2c]] | ||
[[File:Screen shot 2012-02-06 at 9.55.40 AM.png|thumb|center|600px|Both Genlisea and Utricularia are pseudoassemblies against the tomato genome. Compared to syntenic tomato genomic regions. http://genomevolution.org/r/4h43]] | |||
[[File:Screen shot 2012-02-06 at 3.07.02 PM.png|thumb|center|600px|Both Genlisea and Utricularia are pseudoassemblies against the tomato genome. Compared to syntenic tomato genomic regions. '''Note: All HSPs drawn on top''' http://genomevolution.org/r/4h6p]] | |||
==Sytnenic dotplots== | |||
[[File:Screen Shot 2012-02-06 at 6.26.10 AM.png|thumb|left|600px|SynMap analysis of pseudoassembly of Utricularia's contigs (based on synteny to tomato; y-axis) to tomato (x-axis). Results may be regenerated at http://genomevolution.org/r/4h1z]] | |||
Latest revision as of 15:47, 13 February 2012
Overview
Pseudoassemblies for both Utricularia and Genlisea are generated in SynMap using the Syntenic Path Assembly to the tomato genome.
High resolution analyses


Sytnenic dotplots
