Duckweed v. Duckweed: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
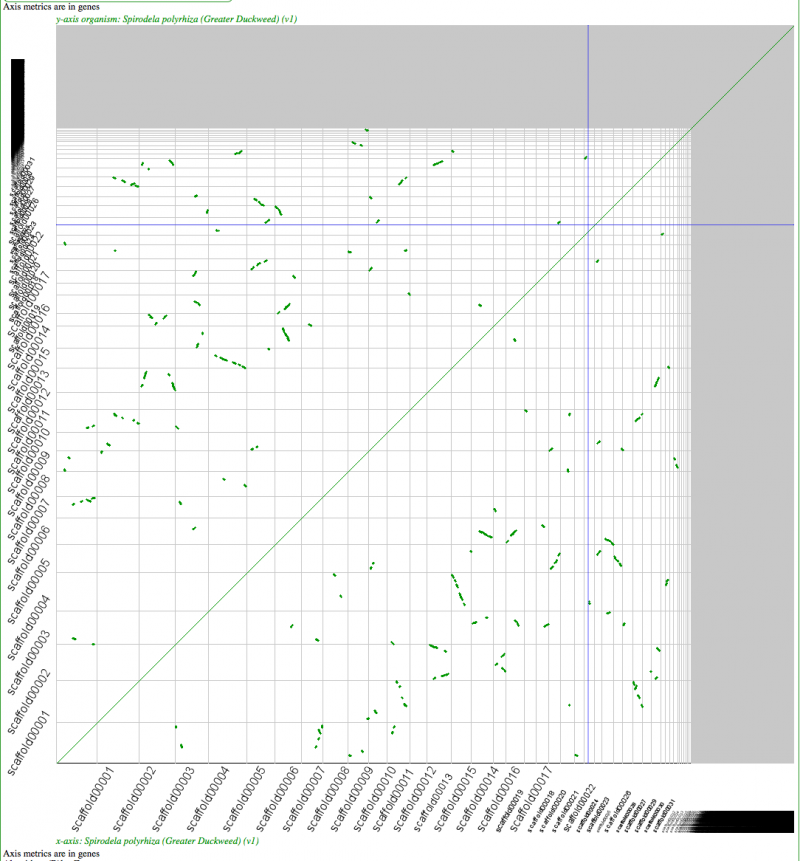
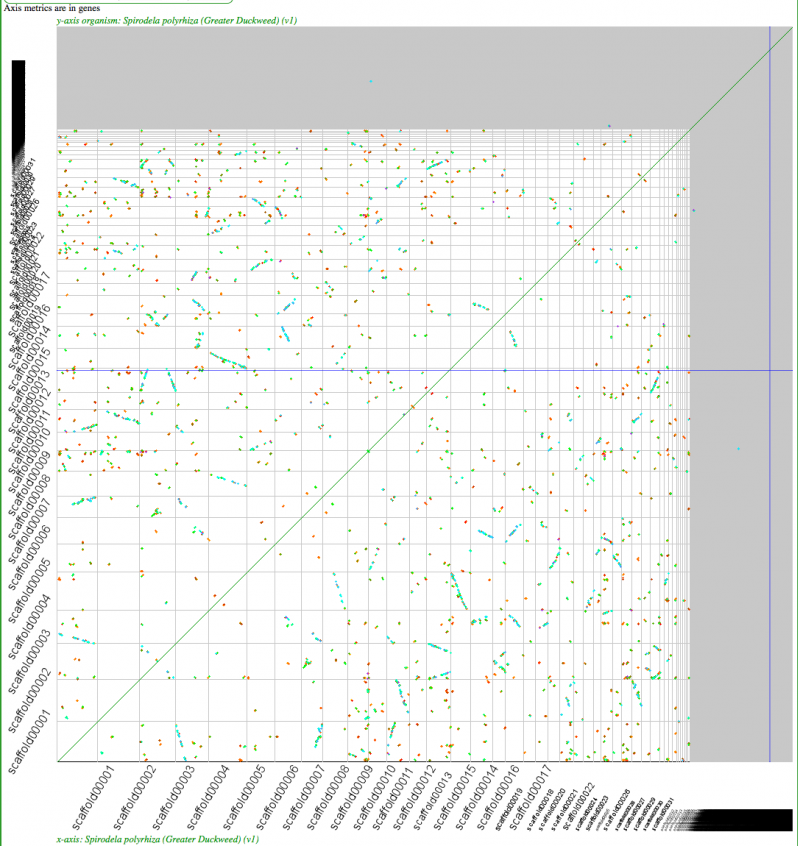
Self-self sytnenic analyses of duckweed has strong evidence for at least one whole genome duplication event. In addition, this may be a hexaploidy event or two sequential tetraploidy events (most likely two tetraploidy events). The [[Ks]] value estimates of syntenic gene pairs, which may be used to differentiate sequential whole genome duplication events (as seen in [[Syntenic comparison of Arabidopsis thaliana and Arabidopsis lyrata | Arabidopsis]]), does not work with Duckweed. This is likely due to the [[Plant genome CDS GC content | high GC content of its coding sequence]]. If duckweed has undergone a recent [[GC content shift]], then all synonymous substitution rate estimates are suspect as not being neutral. As such, Ks values does not yield a reliable metric for differentiating two sequential tetraploidy events in duckweed. | |||
[[File:Screen Shot 2012-03-21 at 6.40.51 PM.png|thumb|800px|center|Syntenic dotplot of self-self with Duckweed. Minimum of 5 genes to call a region syntenic. Results may be regenerated at: http://genomevolution.org/r/4mtn]] | [[File:Screen Shot 2012-03-21 at 6.40.51 PM.png|thumb|800px|center|Syntenic dotplot of self-self with Duckweed. Minimum of 5 genes to call a region syntenic. Results may be regenerated at: http://genomevolution.org/r/4mtn]] | ||
[[File:Screen Shot 2012-03-21 at 7.30.19 PM.png|thumb|800px|center|Syntenic dotplot of self-self with Duckweed. Minimum of 3 genes to call a region syntenic. Results may be regenerated at: http://genomevolution.org/r/4mut]] | [[File:Screen Shot 2012-03-21 at 7.30.19 PM.png|thumb|800px|center|Syntenic dotplot of self-self with Duckweed. Minimum of 3 genes to call a region syntenic. Note: While the synonymous mutation rate calculations are highly suspect due to the high-GC content of this genome and may not differentiate between sequential tetraploidy events, Ks values do differentiate "real" syntenic gene pairs from noise. Results may be regenerated at: http://genomevolution.org/r/4mut]] | ||
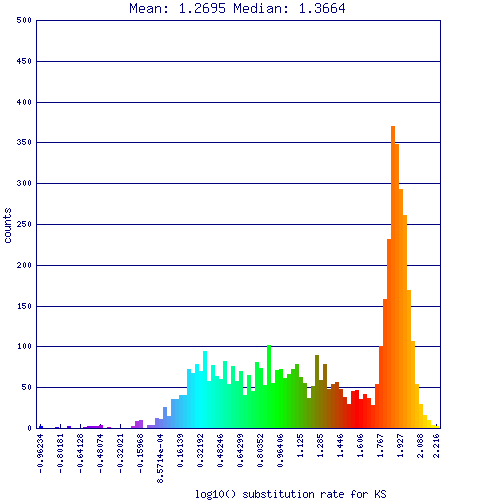
[[File:Master 16886 16886.CDS-CDS.lastz.dag.all.go D20 g10 A3.aligncoords.gcoords ct0.w1000.gene.s.ks.sr.cs1.csoS.log.nsd.hist.png||thumb|600px|center|Ks values for synonymous mutations for syntenic gene pairs identified in self-self comparison of duckweed. These are log10 transformed values, and are rather high for Ks values. The large peak on the right (red-orange) have a Ks value of ~100 substitutions per synonymous site, which is beyond what CODEML can reliably estimate. These values represent noise in the analysis due to the loose parameters used for detecting synteny and false positive syntenic gene pairs were identified.]] | |||
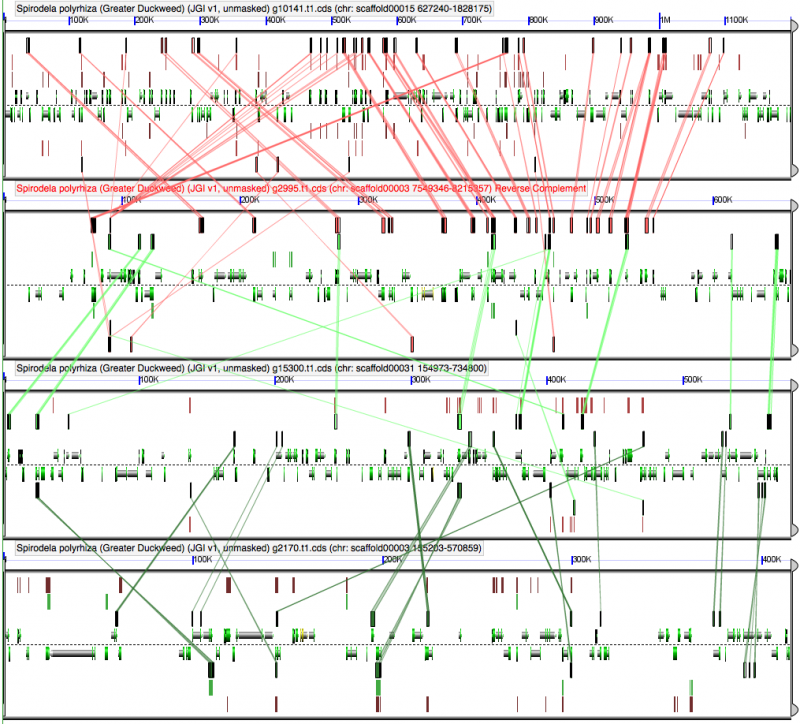
[[File:Screen | [[File:Screen shot 2012-09-28 at 8.56.42 AM.png |thumb|800px|center|[[GEvo]] analysis of 4x duckweed. All regions are syntenic to one another. There is not strong evidence to differentiate two sequential tetraploidies based on have two pairs of regions share more genes with each other than across pairs. Results may be regenerated at: http://genomevolution.org/r/5cvs]] | ||
Latest revision as of 15:59, 28 September 2012
Self-self sytnenic analyses of duckweed has strong evidence for at least one whole genome duplication event. In addition, this may be a hexaploidy event or two sequential tetraploidy events (most likely two tetraploidy events). The Ks value estimates of syntenic gene pairs, which may be used to differentiate sequential whole genome duplication events (as seen in Arabidopsis), does not work with Duckweed. This is likely due to the high GC content of its coding sequence. If duckweed has undergone a recent GC content shift, then all synonymous substitution rate estimates are suspect as not being neutral. As such, Ks values does not yield a reliable metric for differentiating two sequential tetraploidy events in duckweed.