Nelumbo v. Vitis: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
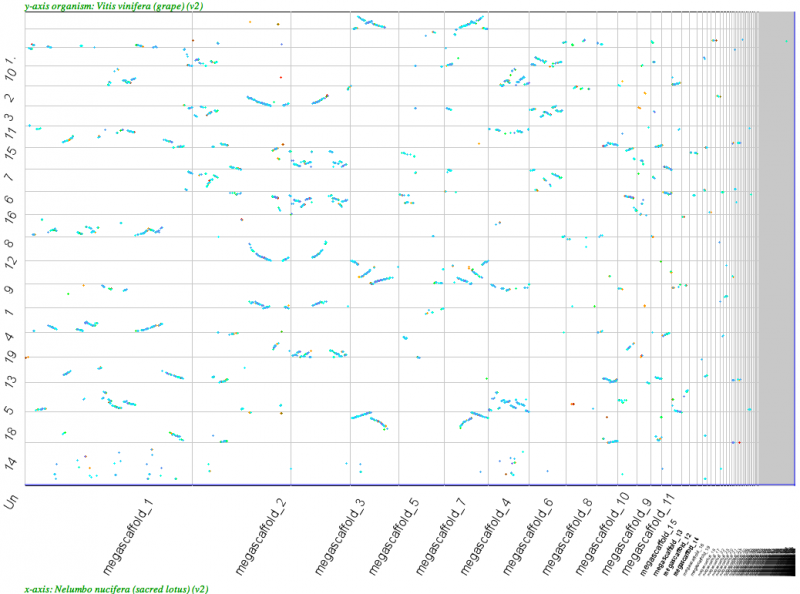
[[File:Screen Shot 2012-03-21 at 1.15.17 PM.png|thumb|center|800px|[[Syntenic dotplot]] of Nelumbo megascaffolds (x-axis) v. Vitus (y-axis). Note the 2xNelumbo to 3xVitus syntenic relationship. Results may be regenerated: http://genomevolution.org/r/4pa5]] | [[File:Screen Shot 2012-03-21 at 1.15.17 PM.png|thumb|center|800px|[[Syntenic dotplot]] of Nelumbo megascaffolds (x-axis) v. Vitus (y-axis). Note the 2xNelumbo to 3xVitus syntenic relationship. Results may be regenerated: http://genomevolution.org/r/4pa5]] | ||
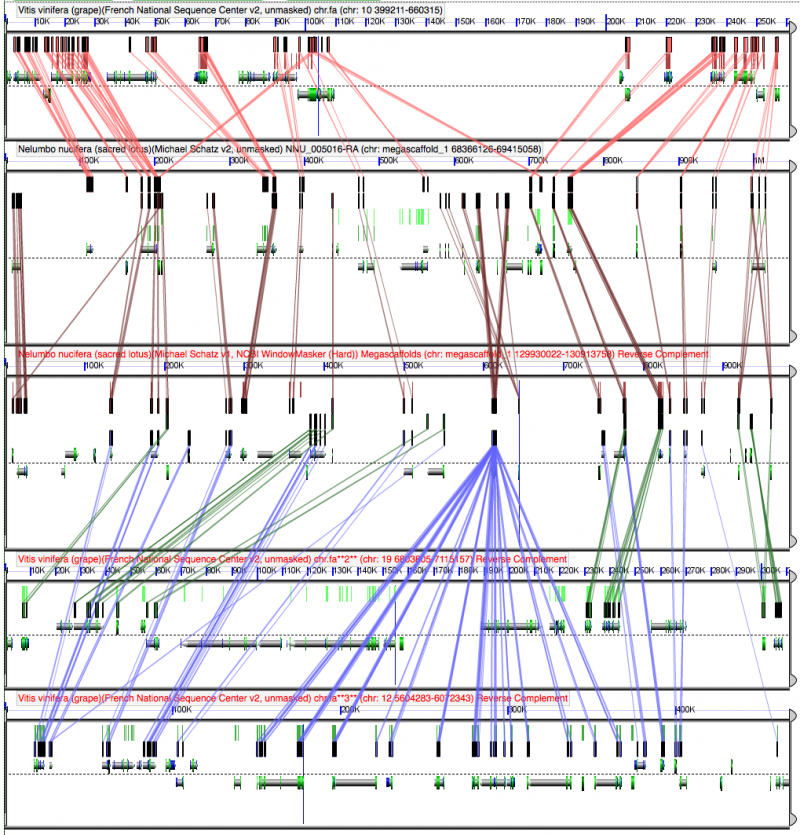
[[File:Screen | [[File:Screen Shot 2012-04-14 at 7.50.47 AM.png|thumb|center|800px|[[GEvo]] high resolution analysis of syntenic regions of Nelumbo nucifera (Nn) and Vitis vinifera (Vv). There are two and three syntenic regions respectively. Colored boxes and lines connect regions of sequence similarity for protein coding sequences. Note their collinear arrangement which is used to infer synteny and that all the regions are differentially fractionated with respect to one another which is evidence for independent polyploidy events ([[tetraploidy]] in the case of Nn and [[hexaploidy]] in the case of Vv. Results may be regenerated at http://genomevolution.org/r/4pt1. Detailed explanation of the figure graphics and how to use GEvo are found at http://genomevolution.org/wiki/index.php?title=GEvo.]] | ||
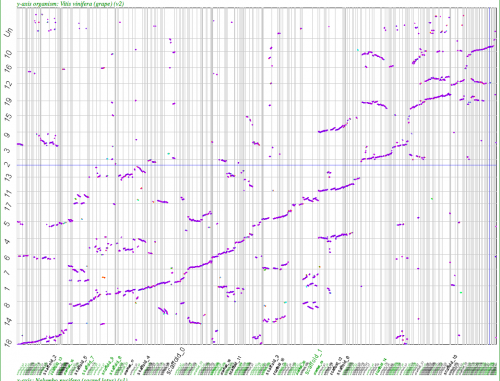
[[File:Screen shot 2012-02-27 at 2.01.45 PM.png|thumb|center|800px|Syntenic dotplot of Nelumbo v. Vitis. Note the 2xNelumbo to 3xVitis syntenic relationship. Syntenic gene pairs colored by Ks values (all are approximately the same). Evidence that duplications are independent of one another. Results may be regenerated: http://genomevolution.org/r/4kei]] | [[File:Screen shot 2012-02-27 at 2.01.45 PM.png|thumb|center|800px|Syntenic dotplot of Nelumbo v. Vitis. Note the 2xNelumbo to 3xVitis syntenic relationship. Syntenic gene pairs colored by Ks values (all are approximately the same). Evidence that duplications are independent of one another. Results may be regenerated: http://genomevolution.org/r/4kei]] | ||
Latest revision as of 15:04, 14 April 2012