Horizontal Gene Transfer: Difference between revisions
No edit summary |
No edit summary |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[Image:K12-HGT.png|thumb|right|600px|A prophage is easily seen in the genome of Escherichia coli K12 MG1655 by its different in gene structure and "red" colored coding sequence representing a high AT content | [[Image:K12-HGT.png|thumb|right|600px|A prophage is easily seen in the genome of Escherichia coli K12 MG1655 by its different in gene structure and "red" colored coding sequence representing a high AT content in the wobble position.]] | ||
One method for detecting horizontal gene transfer events is to find genes that use different codons than the rest of the genome. This is also reflected in the underlying GC content of genes because GC content is correlated with codon usage, which in turn is associated their encoded amino acids. In other words, GC rich genes use GC rich codons. This bias in GC content is often reflected in the wobble position of codons so that GC rich genes use codons that end in G or C. For this example, a cryptic prophage is easily visualized in the genome of Escherichia coli K12 substrain MG1655 by looking for genes that have a different percentage of GC content in the wobble position as compared to the neighboring genomic regions. | |||
To do this type of analysis with CoGe: | |||
#Find an organism of interest using [[GenomeView]] | |||
#Turn on the visualization layer for coloring coding sequence by the percent GC in their wobble positions. | |||
##This is done by pressing the "+" button in the upper right-hand side of [[GenomeView]] and selecting "Wobble GC Usage". | |||
#Now navigate along the genome and look for genes that are different in color than most of the other genes. | |||
The color scheme for the wobble GC usage is: | |||
*green if high GC content in the wobble position | |||
*red if high AT content in the wobble position | |||
*yellow if there is an equal amount of AT and GC in the wobble position | |||
The majority of coding sequence in K12 have a yellow-green color (showing a GC content slightly greater than 50%), the some of the phage's coding sequence is heavily biased towards a high AT content in the wobble position. This results in those coding sequences being colored red. This is further seen by turning on the "Other Feature" visualization layer where a misc_feature that annotates the prophage is shown by a large orange block. This view can be recreated at: http://synteny.cnr.berkeley.edu/CoGe/GenomeView.pl?z=5&x=570000&dsgid=4242&chr=1&dsid=36725&gstid=1 . | |||
Another feature of horizontal genome transfer events, such as this phage integration into a bacterial genome, is that the genome structure is markedly different. While most of this genome is structured into obvious [[operons]], the phage's genome structure is not order as such. | |||
Latest revision as of 18:39, 7 January 2010
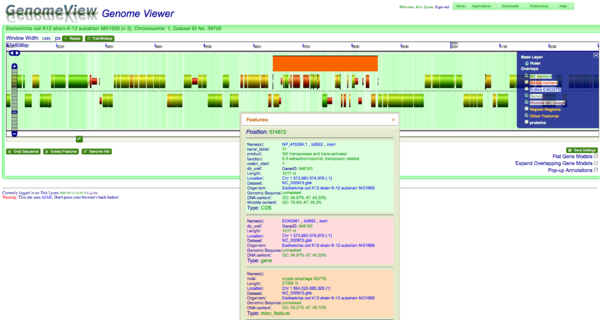
One method for detecting horizontal gene transfer events is to find genes that use different codons than the rest of the genome. This is also reflected in the underlying GC content of genes because GC content is correlated with codon usage, which in turn is associated their encoded amino acids. In other words, GC rich genes use GC rich codons. This bias in GC content is often reflected in the wobble position of codons so that GC rich genes use codons that end in G or C. For this example, a cryptic prophage is easily visualized in the genome of Escherichia coli K12 substrain MG1655 by looking for genes that have a different percentage of GC content in the wobble position as compared to the neighboring genomic regions.
To do this type of analysis with CoGe:
- Find an organism of interest using GenomeView
- Turn on the visualization layer for coloring coding sequence by the percent GC in their wobble positions.
- This is done by pressing the "+" button in the upper right-hand side of GenomeView and selecting "Wobble GC Usage".
- Now navigate along the genome and look for genes that are different in color than most of the other genes.
The color scheme for the wobble GC usage is:
- green if high GC content in the wobble position
- red if high AT content in the wobble position
- yellow if there is an equal amount of AT and GC in the wobble position
The majority of coding sequence in K12 have a yellow-green color (showing a GC content slightly greater than 50%), the some of the phage's coding sequence is heavily biased towards a high AT content in the wobble position. This results in those coding sequences being colored red. This is further seen by turning on the "Other Feature" visualization layer where a misc_feature that annotates the prophage is shown by a large orange block. This view can be recreated at: http://synteny.cnr.berkeley.edu/CoGe/GenomeView.pl?z=5&x=570000&dsgid=4242&chr=1&dsid=36725&gstid=1 .
Another feature of horizontal genome transfer events, such as this phage integration into a bacterial genome, is that the genome structure is markedly different. While most of this genome is structured into obvious operons, the phage's genome structure is not order as such.