LoadBatch: Difference between revisions
No edit summary |
mNo edit summary |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 18: | Line 18: | ||
<span style="color:red">''Note: tarballs must not contain subdirectories.''</span> | <span style="color:red">''Note: tarballs must not contain subdirectories.''</span> | ||
''' | '''The interface allows you to select and retrieve data files located at:''' | ||
*The iPlant Data Store | *The iPlant Data Store | ||
Latest revision as of 18:58, 30 March 2015
LoadBatch provides the ability to conveinently load a set of genomes or experiments in a single operation. To load a set of genomes or experiments using LoadGenome and LoadExperiment would require running the tool for each genome/experiment individually and is very time consuming for large data sets.
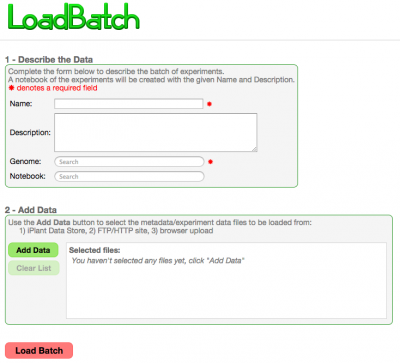
Inputs
Metadata File
A single metadata file that describes the data files contained is required for the load. See the metadata section: Metadata
Data File(s)
Data files can be given individually or together as a compressed tar archive file (ending in .tar.gz, also known as a "tarball").
Valid combinations of input files include:
- tarball of metadata file and data file(s)
- metadata file and tarball of data file(s)
- separate metadata file and data files
Note: tarballs must not contain subdirectories.
The interface allows you to select and retrieve data files located at:
- The iPlant Data Store
- An FTP server
- Your computer (Upload)
Data Formats
For supported genome data file formats, see LoadGenome.
For supported experiment data file formats, see LoadExperiment.