2015 Plant Genome Evolution Workshop: Difference between revisions
| (48 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
===Slides=== | ===Slides=== | ||
* '''Keynote:''' | * '''Keynote (225MB):''' http://de.iplantcollaborative.org/dl/d/7D3BBC8A-1E30-480C-8C3F-0312F6B48F74/2015-PGE-CoGe-Computer-Demo.key | ||
* '''PDF''': | * '''PDF (14MB)''': http://de.iplantcollaborative.org/dl/d/3B7D0F8D-DA26-45E4-ABE3-E4CB634E684E/2015-PGE-CoGe-Computer-Demo.pdf | ||
* '''Powerpoint''': | * '''Powerpoint (210MB)''': http://de.iplantcollaborative.org/dl/d/92B49B0D-68B8-44EF-B10D-E1C430F430BE/2015-PGE-CoGe-Computer-Demo.pptx | ||
===Register an account/Log in=== | ===Register an account/Log in=== | ||
[[File:Screen Shot 2015-08-31 at 4.13.50 PM.png|thumb|300px]] | |||
*Go to: http://user.iplantcollaborative.org | *Go to: http://user.iplantcollaborative.org | ||
**CoGe uses iPlant's Authentication and User Identify Management Service | **CoGe uses iPlant's Authentication and User Identify Management Service | ||
**After clicking on the confirmation link provided in the automated email, your account may take a few minutes to propagate to all of iPlant's Authentication Services. | **After clicking on the confirmation link provided in the automated email, your account may take a few minutes to propagate to all of iPlant's Authentication Services. | ||
*Sign-in (link is in top-right of any CoGe page) | *Sign-in (link is in top-right of any CoGe page) | ||
*'''NOTE:''' This wiki (CoGePedia) uses a different authentication than CoGe! | **'''NOTE:''' This wiki (CoGePedia) uses a different authentication than CoGe! | ||
*Once your are logged in, you have access to "My Profile", CoGe's control page for all of your data and analyses. | |||
===Load your own genome=== | ===Load your own genome=== | ||
[[File:Screen Shot 2015-08-31 at 4.15.56 PM.png|thumb|100px]] | |||
If you are logged into CoGe with your user account, you can add new genomes to CoGe, keep them private, share them with collaborators, and make them fully public. | |||
* Start: | * Start: | ||
** From your user profile page (must be logged into CoGe): https://genomevolution.org/CoGe/User.pl | ** From your user profile page (must be logged into CoGe): https://genomevolution.org/CoGe/User.pl | ||
*** Click "Create"-> "New Genome" | *** Click "Create"-> "New Genome" | ||
*** Go directly to [[LoadGenome]]: https://genomevolution.org/CoGe/LoadGenome.pl | *** Go directly to [[LoadGenome]]: https://genomevolution.org/CoGe/LoadGenome.pl | ||
====Small Genome==== | |||
====Small Genome (E. coli)==== | |||
[[File:Screen Shot 2015-08-31 at 4.23.29 PM.png|thumb|300px]] | |||
* Search for Organism "Escherichia coli K12 strain K-12 substrain MG1655" (just type in "MG1655") | * Search for Organism "Escherichia coli K12 strain K-12 substrain MG1655" (just type in "MG1655") | ||
* Set a version (e.g., "1") | * Set a version (e.g., "1") | ||
| Line 22: | Line 28: | ||
* Source: "CoGe" or "NCBI" | * Source: "CoGe" or "NCBI" | ||
* Leave as "Restricted" | * Leave as "Restricted" | ||
* Press " | * Press '''"Next"''' | ||
* Select "FTP/HTTP" tab | * Select "FTP/HTTP" tab | ||
* Paste in the link below: | * Paste in the link below: | ||
** E. coli genome: http://de.iplantcollaborative.org/dl/d/555B53F9-E738-4951-85A3-421E23804DFA/genome_7112.faa | ** E. coli genome: http://de.iplantcollaborative.org/dl/d/555B53F9-E738-4951-85A3-421E23804DFA/genome_7112.faa | ||
* Press "Get" | * Press '''"Get"''' | ||
* | * Press '''"Next"''' | ||
* Review the data and associated information. | |||
* Press '''"Start Loading"''' | |||
* Note: The length of time it takes to load a genome depends on the load on the database and the number of chromosomes/contigs being loaded. For this example, it should take a minute or two. | * Note: The length of time it takes to load a genome depends on the load on the database and the number of chromosomes/contigs being loaded. For this example, it should take a minute or two. | ||
* Note: When finished, you can select what you want to do next from a drop-down menu: | |||
** Go to [[GenomeInfo]] | |||
** Load Annotations for the genome | |||
** Load Another Genome | |||
====Medium Genome==== | ====Medium Genome (Arabidopsis thaliana)==== | ||
[[File:Screen Shot 2015-08-31 at 4.17.59 PM.png|thumb|300px]] | |||
* Search for Organism "Arabidopsis thaliana Col-0 (thale cress)" (just type in "col-0") | * Search for Organism "Arabidopsis thaliana Col-0 (thale cress)" (just type in "col-0") | ||
* Set a version (e.g., "1") | * Set a version (e.g., "1") | ||
| Line 36: | Line 49: | ||
* Source: "CoGe" or "TAIR" | * Source: "CoGe" or "TAIR" | ||
* Leave as "Restricted" | * Leave as "Restricted" | ||
* Press " | * Press "Next" | ||
* Select "FTP/HTTP" tab | * Select "FTP/HTTP" tab | ||
* Paste in the link below: | * Paste in the link below: | ||
** http://de.iplantcollaborative.org/dl/d/0EF72316-BA37-453A-9297-C07DF9361179/genome_16911.faa | ** Arabidopsis thaliana genome: http://de.iplantcollaborative.org/dl/d/0EF72316-BA37-453A-9297-C07DF9361179/genome_16911.faa | ||
* Press "Get" | * Press "Get" | ||
* | * Press "Next" | ||
* Review the data and associated information. | |||
* Press "Start Loading" | |||
* Note: The length of time it takes to load a genome depends on the load on the database and the number of chromosomes/contigs being loaded. For this example, it should take a minute or two. | * Note: The length of time it takes to load a genome depends on the load on the database and the number of chromosomes/contigs being loaded. For this example, it should take a minute or two. | ||
* Note: When finished, you can select what you want to do next from a drop-down menu: | |||
** Go to [[GenomeInfo]] | |||
** Load Annotations for the genome | |||
** Load Another Genome | |||
===Add Annotations === | |||
If you have structural gene models for your genome, you can integrate them. While many tools can use the full genome, some tools (and some features) require having structural gene models (e.g., CDS). | |||
=== | ====Small genome (E. coli)==== | ||
* Go to GenomeInfo by pressing | [[File:Screen Shot 2015-08-31 at 4.25.54 PM.png|thumb|300px]] | ||
[[File:Screen Shot 2015-08-31 at 4.30.44 PM.png|thumb|300px]] | |||
* Go to [[LoadAnnotation]] | |||
** From [[GenomeInfo]] by pressing "Load Gene Annotations" | |||
** Linked from [[LoadGenome]] | |||
* The genome should automatically be loaded in LoadAnnotation | |||
* Set a version (e.g., "1") | * Set a version (e.g., "1") | ||
* Source: "CoGe" or "NCBI" | * Source: "CoGe" or "NCBI" | ||
* Press '''"Next"''' | |||
* Press " | |||
* Select "FTP/HTTP" Tab | * Select "FTP/HTTP" Tab | ||
* Paste in link below: | * Paste in link below: | ||
** E. coli annotations: http://de.iplantcollaborative.org/dl/d/C1ACC5C3-1E0B-409B-8A91-C969943F41F8/7122-structural-annotations.gff | ** E. coli annotations: http://de.iplantcollaborative.org/dl/d/C1ACC5C3-1E0B-409B-8A91-C969943F41F8/7122-structural-annotations.gff | ||
** Press "Get" | ** Press '''"Get"''' | ||
** | * Press '''"Next"''' | ||
* Review the data and associated information. | |||
* Press '''"Start Loading"''' | |||
* When the load is finished, pressing "Genome View" will launch the genome viewer (JBrowse). | * When the load is finished, pressing "Genome View" will launch the genome viewer (JBrowse). | ||
* '''Note''': The length of time it takes to load annotations depends on the load on the database and the number of annotations being loaded. For this example (and no load on the server), it should take ~ 3-5 minutes. | * '''Note''': The length of time it takes to load annotations depends on the load on the database and the number of annotations being loaded. For this example (and no load on the server), it should take ~ 3-5 minutes. | ||
=== | ====Large genome (Arabidopsis thaliana) ==== | ||
* | * Go to [[LoadAnnotation]] | ||
** | ** From [[GenomeInfo]] by pressing "Load Gene Annotations" | ||
** | ** Linked from [[LoadGenome]] | ||
* | * The genome should automatically be loaded in LoadAnnotation | ||
** | * Set a version (e.g., "1") | ||
** | * Source: "CoGe" or "TAIR" | ||
* | * Press '''"Next"''' | ||
** | * Select "FTP/HTTP" Tab | ||
* | * Paste in link below: | ||
** Arabidopsis thaliana annotations: http://de.iplantcollaborative.org/dl/d/83E2EEAF-68E7-48CD-85E3-7F06CFC81D16/Arabidopsis_thaliana_Col-0_thale_cress_annos1-cds0-id_typename-nu1-upa1-add_chr0.gid16911.gff | |||
** Press '''"Get"''' | |||
* Press '''"Next"''' | |||
* Review the data and associated information. | |||
* Press '''"Start Loading"''' | |||
* When the load is finished, pressing "Genome View" will launch the genome viewer (JBrowse). | |||
* '''Note''': The length of time it takes to load annotations depends on the load on the database and the number of annotations being loaded. For this example (and no load on the server), it should take ~ 10-20 minutes. | |||
===Data Management=== | |||
[[File:Screen Shot 2015-08-31 at 4.19.23 PM.png|thumb|300px]] | |||
CoGe lets you share private data with other users. | |||
* Go to your User Profile page: https://genomevolution.org/CoGe/User.pl | * Go to your User Profile page: https://genomevolution.org/CoGe/User.pl | ||
* Select a genome by clicking on it. | * Select a genome by clicking on it. | ||
| Line 83: | Line 117: | ||
* You can view genomes (and other data) that has been shared with you by clicking on "Shared with me" in the menu on the left. | * You can view genomes (and other data) that has been shared with you by clicking on "Shared with me" in the menu on the left. | ||
=== | ===Adding Experimental Data=== | ||
[[File:Screen Shot 2015-08-31 at 4.32.26 PM.png|thumb|300px]] | |||
[[File:Screen Shot 2015-08-31 at 4.38.02 PM.png|thumb|300px]] | |||
[[File:Screen Shot 2015-08-31 at 4.39.38 PM.png|thumb|300px]] | |||
[[EPIC-CoGe]] is an extension to CoGe that lets you add any type of functional genomics and diversity data sets to CoGe. | |||
* Go to the detailed view for your E. Coli genome from the User Profile Page: https://genomevolution.org/CoGe/User.pl | |||
* Press "Load Experiment" | |||
* '''Note:''' This exercise will be loading expression data, variant data, and read alignment data. You can name your experiments appropriately | |||
* Add a name (e.g., "Expression: Treatment 1") | |||
* Add a description (e.g., "Generated by heat stress and RNASeq protocol 11B") | |||
* Add a version (e.g., "1") | |||
* Add a source (e.g., "iPlant") | |||
* Leave restricted | |||
* Press '''Next''' | |||
* Select the "FTP/HTTP" tab | |||
* Paste in one of the following (based on what you've named your experiment) | |||
** E. coli expression data: http://de.iplantcollaborative.org/dl/d/F1EE1A86-F91A-46E7-92D6-A25EE78E3DA3/7112-lo.csv | |||
** E. coli variant data: http://de.iplantcollaborative.org/dl/d/B4040AC9-74A7-4E3E-B074-F23E1D70823E/7112-demo.vcf.gz | |||
** E. coli bam read alignment data: http://de.iplantcollaborative.org/dl/d/D0FC54E4-6DA6-4604-BE19-81F1D33B5350/7112-alignment.bam | |||
* Press "Get" | |||
* Press '''Next''' | |||
* CoGe should automatically detect the type of file, but you can always set it. The supported file types are available [https://genomevolution.org/wiki/index.php/LoadExperiment#Data_Formats_and_Track_Types here] | |||
* Press '''Next''' | |||
* The options presented depend on the detected file type. For these dataset, you can skip this step. | |||
* Press '''Next''' | |||
* Review the data and associated information. | |||
* Press '''"Start Loading"''' to start the loading the data. | |||
* When finished, there is a link to the experiment you loaded. From there, you can get detailed information on the experiment (and modify what you've entered) in [[ExperimentView]] | |||
* From ExperimentView, you can launch the genome browser by pressing "View" | |||
[[File:Screen Shot 2015-08-31 at 4.49.43 PM.png|thumb|300px]] | |||
[[File:Screen Shot 2015-08-31 at 4.51.19 PM.png|thumb|300px]] | |||
===Visualizing data you just loaded=== | |||
* From your profile page, double click on the genome (or experiment) you loaded | |||
* Press "Browse". This will launch CoGe's implementation of JBrowse: [[GenomeView]] | |||
* You can select tracks to visualize in the menu on the right | |||
===RNASeq Processing=== | |||
[[File:Screen Shot 2015-08-31 at 4.11.52 PM.png|thumb|300px]] | |||
EPIC-CoGe has a variety of pipelines for processing FastQ data. Its [[Expression Analysis Pipeline]] will clean your reads, map them, quantify their abundance to position in the genome, and quantify their abundance per transcript as FPKM values. | |||
* '''Note:''' You can add private experimental data to public genomes (Mix and match public and private data) | |||
* Go to your User Profile page: https://genomevolution.org/coge/User.pl | |||
* Select "Create" -> "New Experiment" | |||
* '''--OR--''' | |||
** You can go to the [[GenomeInfo]] page for Arabisopsis thaliana (https://genomevolution.org/coge/GenomeInfo.pl?gid=16911) and '''Load Experiment''' | |||
** This will send you to [[LoadExperiment]] with the genome field pre-populated | |||
* Add experiment name: (e.g., "RNASeq-test") | |||
* Add description (optional) | |||
* Add version (1) | |||
* Add source (e.g., "coge") | |||
* Keep restricted | |||
* Search for "Col-0" | |||
** Make sure to select the version with genome ID 16911 | |||
* Press '''"Next"''' | |||
* Select the "FTP/HTTP" tab | |||
* Copy in the following link: | |||
** Arabidopsis RNA-Seq data: http://de.iplantcollaborative.org/dl/d/A7F5E57F-E776-46E0-9672-59264E663F8A/test_rna_seq_data_0.17M_reads.fastq | |||
* Press "Get" | |||
* It will automatically detect that it is a fastq file based on the file name extension | |||
* Press '''"Next"''' | |||
* Leave the aligner set to "GSNAP" which is faster than Bowtie2 | |||
* Leave read type to "single-end" | |||
* To use the [[Expression Analysis Pipeline]], select the check-box next to "Enable" | |||
** For parameters on this test dataset, use the "CoGe Basic" method and set the minimum read depth to "1" (the example data is a subsample of a fastq file and contains 170,000 reads) | |||
* You can tell CoGe to automatically add the newly loaded data to a notebook by selecting the check-box next to "Add results to notebook" | |||
* You can tell CoGe to send you and email when the analysis is done by selecting the check-box next to "Send email when finished" | |||
* Press '''"Next"''' | |||
* Review the data and associated information. | |||
* Press '''"Start Loading"''' to start the analysis. | |||
** '''Note:''' Once the analysis starts running, you can close your browser. | |||
** '''Note:''' You can check on the status of your analysis by clicking on "Data Loading" under "Activity" in your Profile Page | |||
* '''Note:''' This Fastq file is relatively small and the whole pipeline takes around 2-3 minutes to complete | |||
* When finished, Load Experiment will create a notebook (if selected) with three experiments as well provide links to each experiment. | |||
** One for the BAM file (alignment) | |||
** One for reads mapped to nucleotide positions in the genome (read depth) | |||
** One for reads normalized to transcripts (FPKM) | |||
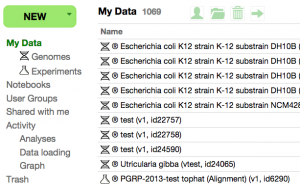
* You can find your newly loaded experiments by clicking on "Experiments" under "My Data" | |||
** From there, you can double click on the experiment to get information about it along with links to visualize the data in JBrowse | |||
===SNP and Variant Detection=== | |||
[[File:Screen Shot 2015-08-31 at 4.54.30 PM.png|thumb|300px]] | |||
You can identify SNPs/variants in CoGe by following the RNASeq processing tutorial, but selecting to enable SNP Identification | |||
*Details on the SNP processing pipeline are found [https://genomevolution.org/wiki/index.php/LoadExperiment#SNP_Identification_Pipeline here] | |||
===Find and visualize a genome=== | |||
* Find a genome using [[OrganismView]] | |||
** Search for E. coli MG1655 my typing "MG1655" in the search box | |||
** E. coli MG1655: https://genomevolution.org/CoGe/OrganismView.pl?oid=24290 | |||
* Detailed Genome Information in [[GenomeInfo]] | |||
** Follow the link to GenomeInfo under "Genome Information" | |||
** E. coli MG1655: https://genomevolution.org/CoGe/GenomeInfo.pl?gid=4242 | |||
* Visualize Genome in [[GenomeView]] | |||
** View the genome in the genome browser by pressing the "View" button | |||
** E. coli MG1655: https://genomevolution.org/CoGe/GenomeView.pl?gid=4242 | |||
===Comparative Genomic Analyses=== | |||
[[File:Screen Shot 2015-08-31 at 4.58.37 PM.png|thumb|300px]] | |||
* Get the detailed view of your genome ([[GenomeInfo]]) | * Get the detailed view of your genome ([[GenomeInfo]]) | ||
* Under "Tools" and next to "Analyze", click on the link for "[[SynMap]]" | * Under "Tools" and next to "Analyze", click on the link for "[[SynMap]]" | ||
| Line 103: | Line 236: | ||
* Your previously loaded data can be viewed by clicking "Data loading". Clicking on a previously loaded data will open the detailed view for those data. | * Your previously loaded data can be viewed by clicking "Data loading". Clicking on a previously loaded data will open the detailed view for those data. | ||
===Comparative Genomics=== | ===Comparative Genomics=== | ||
[[File:CoGe SynMap2.png|thumb|300px]] | |||
* Human-Chimp whole genome: http://genomevolution.org/r/d76i | * Human-Chimp whole genome: http://genomevolution.org/r/d76i | ||
** Go to [[SynMap]]: https://genomevolution.org/CoGe/SynMap.pl | ** Go to [[SynMap]]: https://genomevolution.org/CoGe/SynMap.pl | ||
| Line 165: | Line 275: | ||
*** When the results are returned, there is a link to analyze the identified syntenic regions with GEvo to confirm synteny. | *** When the results are returned, there is a link to analyze the identified syntenic regions with GEvo to confirm synteny. | ||
** Microsynteny analysis: http://genomevolution.org/r/d778 | ** Microsynteny analysis: http://genomevolution.org/r/d778 | ||
===Data files reference=== | ===Data files reference=== | ||
| Line 200: | Line 282: | ||
* E. coli variant data: http://de.iplantcollaborative.org/dl/d/B4040AC9-74A7-4E3E-B074-F23E1D70823E/7112-demo.vcf.gz | * E. coli variant data: http://de.iplantcollaborative.org/dl/d/B4040AC9-74A7-4E3E-B074-F23E1D70823E/7112-demo.vcf.gz | ||
* E. coli bam alignment data: http://de.iplantcollaborative.org/dl/d/D0FC54E4-6DA6-4604-BE19-81F1D33B5350/7112-alignment.bam | * E. coli bam alignment data: http://de.iplantcollaborative.org/dl/d/D0FC54E4-6DA6-4604-BE19-81F1D33B5350/7112-alignment.bam | ||
* Arabidopsis thaliana genome: http://de.iplantcollaborative.org/dl/d/0EF72316-BA37-453A-9297-C07DF9361179/genome_16911.faa | |||
* Arabidopsis thaliana annotations: http://de.iplantcollaborative.org/dl/d/83E2EEAF-68E7-48CD-85E3-7F06CFC81D16/Arabidopsis_thaliana_Col-0_thale_cress_annos1-cds0-id_typename-nu1-upa1-add_chr0.gid16911.gff | |||
* Arabidopsis RNA-Seq data: http://de.iplantcollaborative.org/dl/d/A7F5E57F-E776-46E0-9672-59264E663F8A/test_rna_seq_data_0.17M_reads.fastq | * Arabidopsis RNA-Seq data: http://de.iplantcollaborative.org/dl/d/A7F5E57F-E776-46E0-9672-59264E663F8A/test_rna_seq_data_0.17M_reads.fastq | ||
Latest revision as of 18:03, 2 September 2015
Slides
- Keynote (225MB): http://de.iplantcollaborative.org/dl/d/7D3BBC8A-1E30-480C-8C3F-0312F6B48F74/2015-PGE-CoGe-Computer-Demo.key
- PDF (14MB): http://de.iplantcollaborative.org/dl/d/3B7D0F8D-DA26-45E4-ABE3-E4CB634E684E/2015-PGE-CoGe-Computer-Demo.pdf
- Powerpoint (210MB): http://de.iplantcollaborative.org/dl/d/92B49B0D-68B8-44EF-B10D-E1C430F430BE/2015-PGE-CoGe-Computer-Demo.pptx
Register an account/Log in
- Go to: http://user.iplantcollaborative.org
- CoGe uses iPlant's Authentication and User Identify Management Service
- After clicking on the confirmation link provided in the automated email, your account may take a few minutes to propagate to all of iPlant's Authentication Services.
- Sign-in (link is in top-right of any CoGe page)
- NOTE: This wiki (CoGePedia) uses a different authentication than CoGe!
- Once your are logged in, you have access to "My Profile", CoGe's control page for all of your data and analyses.
Load your own genome
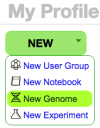
If you are logged into CoGe with your user account, you can add new genomes to CoGe, keep them private, share them with collaborators, and make them fully public.
- Start:
- From your user profile page (must be logged into CoGe): https://genomevolution.org/CoGe/User.pl
- Click "Create"-> "New Genome"
- Go directly to LoadGenome: https://genomevolution.org/CoGe/LoadGenome.pl
- From your user profile page (must be logged into CoGe): https://genomevolution.org/CoGe/User.pl
Small Genome (E. coli)

- Search for Organism "Escherichia coli K12 strain K-12 substrain MG1655" (just type in "MG1655")
- Set a version (e.g., "1")
- Leave "Type:" as "unmasked"
- Source: "CoGe" or "NCBI"
- Leave as "Restricted"
- Press "Next"
- Select "FTP/HTTP" tab
- Paste in the link below:
- Press "Get"
- Press "Next"
- Review the data and associated information.
- Press "Start Loading"
- Note: The length of time it takes to load a genome depends on the load on the database and the number of chromosomes/contigs being loaded. For this example, it should take a minute or two.
- Note: When finished, you can select what you want to do next from a drop-down menu:
- Go to GenomeInfo
- Load Annotations for the genome
- Load Another Genome
Medium Genome (Arabidopsis thaliana)
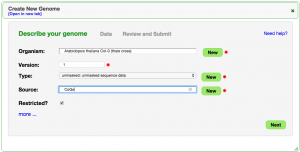
- Search for Organism "Arabidopsis thaliana Col-0 (thale cress)" (just type in "col-0")
- Set a version (e.g., "1")
- Leave "Type:" as "unmasked"
- Source: "CoGe" or "TAIR"
- Leave as "Restricted"
- Press "Next"
- Select "FTP/HTTP" tab
- Paste in the link below:
- Arabidopsis thaliana genome: http://de.iplantcollaborative.org/dl/d/0EF72316-BA37-453A-9297-C07DF9361179/genome_16911.faa
- Press "Get"
- Press "Next"
- Review the data and associated information.
- Press "Start Loading"
- Note: The length of time it takes to load a genome depends on the load on the database and the number of chromosomes/contigs being loaded. For this example, it should take a minute or two.
- Note: When finished, you can select what you want to do next from a drop-down menu:
- Go to GenomeInfo
- Load Annotations for the genome
- Load Another Genome
Add Annotations
If you have structural gene models for your genome, you can integrate them. While many tools can use the full genome, some tools (and some features) require having structural gene models (e.g., CDS).
Small genome (E. coli)

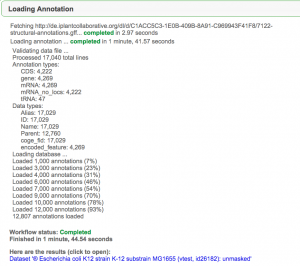
- Go to LoadAnnotation
- From GenomeInfo by pressing "Load Gene Annotations"
- Linked from LoadGenome
- The genome should automatically be loaded in LoadAnnotation
- Set a version (e.g., "1")
- Source: "CoGe" or "NCBI"
- Press "Next"
- Select "FTP/HTTP" Tab
- Paste in link below:
- E. coli annotations: http://de.iplantcollaborative.org/dl/d/C1ACC5C3-1E0B-409B-8A91-C969943F41F8/7122-structural-annotations.gff
- Press "Get"
- Press "Next"
- Review the data and associated information.
- Press "Start Loading"
- When the load is finished, pressing "Genome View" will launch the genome viewer (JBrowse).
- Note: The length of time it takes to load annotations depends on the load on the database and the number of annotations being loaded. For this example (and no load on the server), it should take ~ 3-5 minutes.
Large genome (Arabidopsis thaliana)
- Go to LoadAnnotation
- From GenomeInfo by pressing "Load Gene Annotations"
- Linked from LoadGenome
- The genome should automatically be loaded in LoadAnnotation
- Set a version (e.g., "1")
- Source: "CoGe" or "TAIR"
- Press "Next"
- Select "FTP/HTTP" Tab
- Paste in link below:
- Arabidopsis thaliana annotations: http://de.iplantcollaborative.org/dl/d/83E2EEAF-68E7-48CD-85E3-7F06CFC81D16/Arabidopsis_thaliana_Col-0_thale_cress_annos1-cds0-id_typename-nu1-upa1-add_chr0.gid16911.gff
- Press "Get"
- Press "Next"
- Review the data and associated information.
- Press "Start Loading"
- When the load is finished, pressing "Genome View" will launch the genome viewer (JBrowse).
- Note: The length of time it takes to load annotations depends on the load on the database and the number of annotations being loaded. For this example (and no load on the server), it should take ~ 10-20 minutes.
Data Management
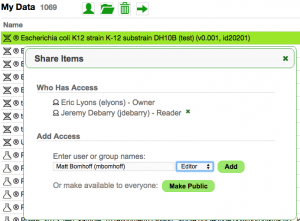
CoGe lets you share private data with other users.
- Go to your User Profile page: https://genomevolution.org/CoGe/User.pl
- Select a genome by clicking on it.
- Information about the genome will appear in the right panel
- You can share a genome by clicking on the person icon
- You can delete the genome by clicking on the trash can
- Double-clicking the genome will open the Detailed View for it (GenomeInfo)
- Share it with the person next to you
- You can view genomes (and other data) that has been shared with you by clicking on "Shared with me" in the menu on the left.
Adding Experimental Data
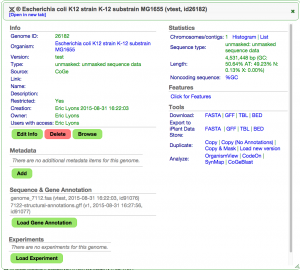
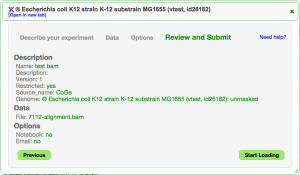
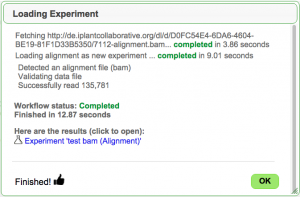
EPIC-CoGe is an extension to CoGe that lets you add any type of functional genomics and diversity data sets to CoGe.
- Go to the detailed view for your E. Coli genome from the User Profile Page: https://genomevolution.org/CoGe/User.pl
- Press "Load Experiment"
- Note: This exercise will be loading expression data, variant data, and read alignment data. You can name your experiments appropriately
- Add a name (e.g., "Expression: Treatment 1")
- Add a description (e.g., "Generated by heat stress and RNASeq protocol 11B")
- Add a version (e.g., "1")
- Add a source (e.g., "iPlant")
- Leave restricted
- Press Next
- Select the "FTP/HTTP" tab
- Paste in one of the following (based on what you've named your experiment)
- E. coli expression data: http://de.iplantcollaborative.org/dl/d/F1EE1A86-F91A-46E7-92D6-A25EE78E3DA3/7112-lo.csv
- E. coli variant data: http://de.iplantcollaborative.org/dl/d/B4040AC9-74A7-4E3E-B074-F23E1D70823E/7112-demo.vcf.gz
- E. coli bam read alignment data: http://de.iplantcollaborative.org/dl/d/D0FC54E4-6DA6-4604-BE19-81F1D33B5350/7112-alignment.bam
- Press "Get"
- Press Next
- CoGe should automatically detect the type of file, but you can always set it. The supported file types are available here
- Press Next
- The options presented depend on the detected file type. For these dataset, you can skip this step.
- Press Next
- Review the data and associated information.
- Press "Start Loading" to start the loading the data.
- When finished, there is a link to the experiment you loaded. From there, you can get detailed information on the experiment (and modify what you've entered) in ExperimentView
- From ExperimentView, you can launch the genome browser by pressing "View"
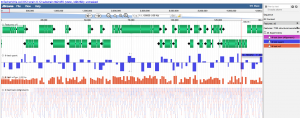
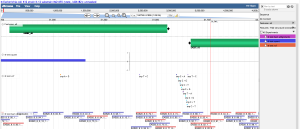
Visualizing data you just loaded
- From your profile page, double click on the genome (or experiment) you loaded
- Press "Browse". This will launch CoGe's implementation of JBrowse: GenomeView
- You can select tracks to visualize in the menu on the right
RNASeq Processing
EPIC-CoGe has a variety of pipelines for processing FastQ data. Its Expression Analysis Pipeline will clean your reads, map them, quantify their abundance to position in the genome, and quantify their abundance per transcript as FPKM values.
- Note: You can add private experimental data to public genomes (Mix and match public and private data)
- Go to your User Profile page: https://genomevolution.org/coge/User.pl
- Select "Create" -> "New Experiment"
- --OR--
- You can go to the GenomeInfo page for Arabisopsis thaliana (https://genomevolution.org/coge/GenomeInfo.pl?gid=16911) and Load Experiment
- This will send you to LoadExperiment with the genome field pre-populated
- Add experiment name: (e.g., "RNASeq-test")
- Add description (optional)
- Add version (1)
- Add source (e.g., "coge")
- Keep restricted
- Search for "Col-0"
- Make sure to select the version with genome ID 16911
- Press "Next"
- Select the "FTP/HTTP" tab
- Copy in the following link:
- Press "Get"
- It will automatically detect that it is a fastq file based on the file name extension
- Press "Next"
- Leave the aligner set to "GSNAP" which is faster than Bowtie2
- Leave read type to "single-end"
- To use the Expression Analysis Pipeline, select the check-box next to "Enable"
- For parameters on this test dataset, use the "CoGe Basic" method and set the minimum read depth to "1" (the example data is a subsample of a fastq file and contains 170,000 reads)
- You can tell CoGe to automatically add the newly loaded data to a notebook by selecting the check-box next to "Add results to notebook"
- You can tell CoGe to send you and email when the analysis is done by selecting the check-box next to "Send email when finished"
- Press "Next"
- Review the data and associated information.
- Press "Start Loading" to start the analysis.
- Note: Once the analysis starts running, you can close your browser.
- Note: You can check on the status of your analysis by clicking on "Data Loading" under "Activity" in your Profile Page
- Note: This Fastq file is relatively small and the whole pipeline takes around 2-3 minutes to complete
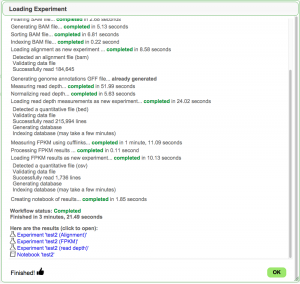
- When finished, Load Experiment will create a notebook (if selected) with three experiments as well provide links to each experiment.
- One for the BAM file (alignment)
- One for reads mapped to nucleotide positions in the genome (read depth)
- One for reads normalized to transcripts (FPKM)
- You can find your newly loaded experiments by clicking on "Experiments" under "My Data"
- From there, you can double click on the experiment to get information about it along with links to visualize the data in JBrowse
SNP and Variant Detection
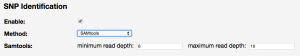
You can identify SNPs/variants in CoGe by following the RNASeq processing tutorial, but selecting to enable SNP Identification
- Details on the SNP processing pipeline are found here
Find and visualize a genome
- Find a genome using OrganismView
- Search for E. coli MG1655 my typing "MG1655" in the search box
- E. coli MG1655: https://genomevolution.org/CoGe/OrganismView.pl?oid=24290
- Detailed Genome Information in GenomeInfo
- Follow the link to GenomeInfo under "Genome Information"
- E. coli MG1655: https://genomevolution.org/CoGe/GenomeInfo.pl?gid=4242
- Visualize Genome in GenomeView
- View the genome in the genome browser by pressing the "View" button
- E. coli MG1655: https://genomevolution.org/CoGe/GenomeView.pl?gid=4242
Comparative Genomic Analyses
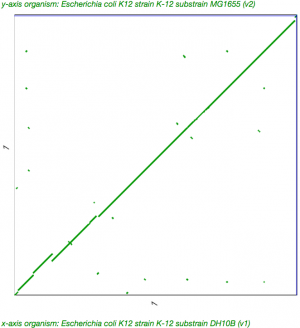
- Get the detailed view of your genome (GenomeInfo)
- Under "Tools" and next to "Analyze", click on the link for "SynMap"
- Your genome will automatically be populated for both genomes
- Search for another E. coli genome by typing "MG1655" into one of the Organism search boxes
- The one auto-select from that search will be perfect for the analysis (ID 4242)
- Scroll to the bottom of the page and press the red button "Generate SynMap"
- When the analysis is finished, press "Go" to see the results
- Click on the dotpot to get a zoomed-in version of the dotplot.
- Scroll onto the green line in the dotplot and double click when the cross-hairs turn red to launch GEvo for microsynteny analysis
- Press "Run GEvo" to run GEvo
- Note: This link will run a SynMap analysis for E. coli K12 substrains MG1655 and DH10B: https://genomevolution.org/r/daqz
Your History and Activity
- If you are logged into CoGe, CoGe will record your activities. These are available for review in your User Profile page: https://genomevolution.org/CoGe/User.pl
- Click on "Activity" in the menu on the left. This will give you an overview of the number of analyses you've run
- Your previously run analyses can be viewed by clicking "Analyses". Clicking on an analysis will re-run it.
- Your previously loaded data can be viewed by clicking "Data loading". Clicking on a previously loaded data will open the detailed view for those data.
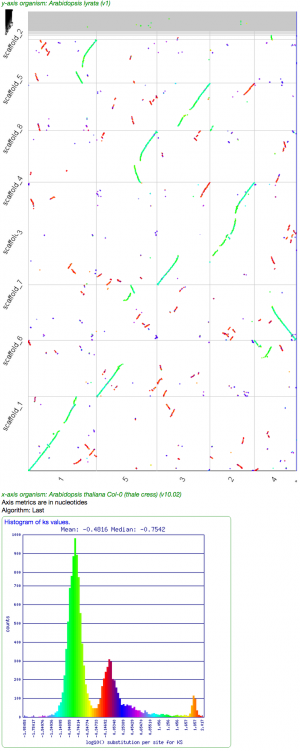
Comparative Genomics
- Human-Chimp whole genome: http://genomevolution.org/r/d76i
- Go to SynMap: https://genomevolution.org/CoGe/SynMap.pl
- Search for "human" and "chimp" in each of the Organism search boxes.
- Select the "Display Options" tab and "Sort Chromosomes" by "Name"
- Press "Generate SynMap"
- Arabidopsis thaliana self-self: http://genomevolution.org/r/d7p5
- Go to SynMap: https://genomevolution.org/CoGe/SynMap.pl
- Search for "Col-0" in both of the Organism search boxes
- Note: many people are making copies of Arabidopsis Col-0 -- select the one with genome ID 16911
- Press "Generate SynMap"
- Arabidopsis thaliana v. Arabidopsis lyrata: http://genomevolution.org/r/d4hi
- Go to SynMap: https://genomevolution.org/CoGe/SynMap.pl
- Search for "Col-0" and "lyrata" in each of the Organism search boxes.
- Press "Generate SynMap"
- Arabidopsis thaliana v. Arabidopsis lyrata (Synonymous values): http://genomevolution.org/r/d7e7
- Go to SynMap: https://genomevolution.org/CoGe/SynMap.pl
- Search for "Col-0" and "lyrata" in each of the Organism search boxes.
- Select the "Analysis Options" tab near the top of the screen
- Under "CodeML" and next to "Calculate syntenic CDS pairs and color dots", select "Synonymous (Ks)"
- Press "Generate SynMap"
- SynFind
- Find a peach gene in the genome browser: https://genomevolution.org/CoGe/GenomeView.pl?chr=scaffold_1&gid=22743&x=10266537&z=6&loc=scaffold_1%3A10266380..10666380&tracks=sequence%2Cfeatures
- Click on a peach gene to get detailed information about it. This includes links to tools in CoGe for downstream analysis
- Send to SynFind by clicking on "SynFind" next to "CoGe Links" : https://genomevolution.org/CoGe/SynFind.pl?fid=461399390
- Note: SynFind is CoGe's tool for identify syntenic regions in any number of other genomes using a gene to anchor your location in a query genome. This tool only works on genomes with CDS annotations.
- Search and add the genome of Arabidopsis thaliana Col-0: http://genomevolution.org/r/d774
- Search for "Col-0" in the Organism search box
- Select the TAIR genome (id 16911)
- Add the Arabidopsis genome by pressing the "Add" button
- Run SynFind by pressing "Run SynFind"
- When the results are returned, there is a link to analyze the identified syntenic regions with GEvo to confirm synteny.
- Microsynteny analysis: http://genomevolution.org/r/d778
Data files reference
- E. coli genome: http://de.iplantcollaborative.org/dl/d/555B53F9-E738-4951-85A3-421E23804DFA/genome_7112.faa
- E. coli annotations: http://de.iplantcollaborative.org/dl/d/C1ACC5C3-1E0B-409B-8A91-C969943F41F8/7122-structural-annotations.gff
- E. coli expression data: http://de.iplantcollaborative.org/dl/d/F1EE1A86-F91A-46E7-92D6-A25EE78E3DA3/7112-lo.csv
- E. coli variant data: http://de.iplantcollaborative.org/dl/d/B4040AC9-74A7-4E3E-B074-F23E1D70823E/7112-demo.vcf.gz
- E. coli bam alignment data: http://de.iplantcollaborative.org/dl/d/D0FC54E4-6DA6-4604-BE19-81F1D33B5350/7112-alignment.bam
- Arabidopsis thaliana genome: http://de.iplantcollaborative.org/dl/d/0EF72316-BA37-453A-9297-C07DF9361179/genome_16911.faa
- Arabidopsis thaliana annotations: http://de.iplantcollaborative.org/dl/d/83E2EEAF-68E7-48CD-85E3-7F06CFC81D16/Arabidopsis_thaliana_Col-0_thale_cress_annos1-cds0-id_typename-nu1-upa1-add_chr0.gid16911.gff
- Arabidopsis RNA-Seq data: http://de.iplantcollaborative.org/dl/d/A7F5E57F-E776-46E0-9672-59264E663F8A/test_rna_seq_data_0.17M_reads.fastq
CoGe Learning Material
- Loading private data in CoGe: http://www.youtube.com/watch?v=JPZ8IrPnh_8
- RNASeq with EPIC-CoGe: https://www.youtube.com/watch?v=3fNyHGB02dM
- Tutorials: https://genomevolution.org/wiki/index.php/Tutorials
- Arabidopsis/Brassica: http://journal.frontiersin.org/Journal/10.3389/fpls.2012.00172/full
- Grasses (maize/sorghum): http://www.maydica.org/articles/56_183.pdf