Maize Sorghum Syntenic dotplot: Difference between revisions
No edit summary |
No edit summary |
||
| Line 9: | Line 9: | ||
# the divergence of the maize and sorghum lineages | # the divergence of the maize and sorghum lineages | ||
# the maize lineage-specific whole genome duplication event | # the maize lineage-specific whole genome duplication event | ||
Each one of these events creates a copy of the genome, and these events can be seen in a [[syntenic dotplot]] between these genomes. A whole genome syntenic dotplot takes two genomes and lays them out end-to-end along each axis. In the figure shown to the right, the sorghum genome is on the x-axis, and the maize genome is on the y-axis. Each black vertical and horizontal line delineates a chromosome. Each gene from those genomes are compared to one another and a dot is drawn at the appropriate x-y coordinate if two genes are similar in sequence, and hence are putative [[homologs]]. These results are then fed into an algorithm to find collinear series of genes. The idea is that if two genomic regions are related to one another through common descent from the same ancestral genomic region, then they will maintain a collinear arrangement of genes from that ancestor. While genomes can change, and genes can move to new genomic positions, this idea can be used to infer that two genomic regions are related through common ancestry (synteny) if there is a collinear arrangement of genes. When such collinear arrangements are detected in this syntenic dotplot, those dots get colored. We call pairs of genes in a collinear arrangement [[syntenic gene pairs]], or [[syntelogs]]. | Each one of these events creates a copy of the genome, and these events can be seen in a [[syntenic dotplot]] between these genomes. A whole genome syntenic dotplot takes two genomes and lays them out end-to-end along each axis. In the figure shown to the right, the sorghum genome is on the x-axis, and the maize genome is on the y-axis. Each black vertical and horizontal line delineates a chromosome. Each gene from those genomes are compared to one another and a dot is drawn at the appropriate x-y coordinate if two genes are similar in sequence, and hence are putative [[homologs]]. These results are then fed into an algorithm to find collinear series of genes. The idea is that if two genomic regions are related to one another through common descent from the same ancestral genomic region, then they will maintain a collinear arrangement of genes from that ancestor. While genomes can change, and genes can move to new genomic positions, this idea can be used to infer that two genomic regions are related through common ancestry ([[synteny]]) if there is a collinear arrangement of genes. When such collinear arrangements are detected in this syntenic dotplot, those dots get colored. We call pairs of genes in a collinear arrangement [[syntenic gene pairs]], or [[syntelogs]]. | ||
Since the whole genome duplication and lineage divergence events that happened in the lineages of maize and sorghum happened at different times, the gene-pairs derived from those events are also of different ages. One way to measure the relative age of a pair of related genes is by estimating their rates of [[synonymous mutations]]. Genes that are more closely related have fewer synonymous changes than genes that are more distantly related. The rate of synonymous change has been measured for each pair of syntelogs identified in the maize-sorghum syntenic dotplot, and colored such that younger syntelogs are colored red, and older syntelogs are colored purple. Looking at the syntenic dotplot, it is now easy to identify red, younger sytnenic regions and purple, older syntenic regions. | Since the whole genome duplication and lineage divergence events that happened in the lineages of maize and sorghum happened at different times, the gene-pairs derived from those events are also of different ages. One way to measure the relative age of a pair of related genes is by estimating their rates of [[synonymous mutations]]. Genes that are more closely related have fewer synonymous changes than genes that are more distantly related. The rate of synonymous change has been measured for each pair of syntelogs identified in the maize-sorghum syntenic dotplot, and colored such that younger syntelogs are colored red, and older syntelogs are colored purple. Looking at the syntenic dotplot, it is now easy to identify red, younger sytnenic regions and purple, older syntenic regions. | ||
Revision as of 19:45, 28 December 2009
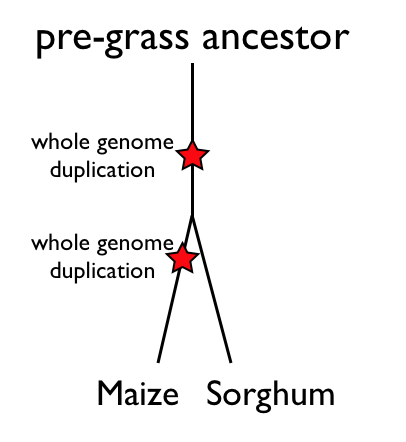

When comparing the genomes of maize and sorghum, there are three events that need to be considered. These events, listed in chronological order, are:
- a whole genome duplication event that is shared among all the grasses
- the divergence of the maize and sorghum lineages
- the maize lineage-specific whole genome duplication event
Each one of these events creates a copy of the genome, and these events can be seen in a syntenic dotplot between these genomes. A whole genome syntenic dotplot takes two genomes and lays them out end-to-end along each axis. In the figure shown to the right, the sorghum genome is on the x-axis, and the maize genome is on the y-axis. Each black vertical and horizontal line delineates a chromosome. Each gene from those genomes are compared to one another and a dot is drawn at the appropriate x-y coordinate if two genes are similar in sequence, and hence are putative homologs. These results are then fed into an algorithm to find collinear series of genes. The idea is that if two genomic regions are related to one another through common descent from the same ancestral genomic region, then they will maintain a collinear arrangement of genes from that ancestor. While genomes can change, and genes can move to new genomic positions, this idea can be used to infer that two genomic regions are related through common ancestry (synteny) if there is a collinear arrangement of genes. When such collinear arrangements are detected in this syntenic dotplot, those dots get colored. We call pairs of genes in a collinear arrangement syntenic gene pairs, or syntelogs.
Since the whole genome duplication and lineage divergence events that happened in the lineages of maize and sorghum happened at different times, the gene-pairs derived from those events are also of different ages. One way to measure the relative age of a pair of related genes is by estimating their rates of synonymous mutations. Genes that are more closely related have fewer synonymous changes than genes that are more distantly related. The rate of synonymous change has been measured for each pair of syntelogs identified in the maize-sorghum syntenic dotplot, and colored such that younger syntelogs are colored red, and older syntelogs are colored purple. Looking at the syntenic dotplot, it is now easy to identify red, younger sytnenic regions and purple, older syntenic regions.
Looking closely at the syntenic dotplot, there is an overlap of these colored lines. This is because a given region of one genome is syntenic to multiple regions in the other genome. Based on the series of events listed above, it is expected that for every region of the sorghum genome, there will be two red lines in maize because maize has had a whole genome duplication event after these lineages diverged. On the other hand, for each region of the maize genome, there will only be one red line in sorghum.
Understanding the purple lines is a bit more complicated, but not by much. This come form the older shared whole genome duplication event. As seen with the red lines, for a given region of sorghum, there should be two purple lines that come from maize's most recent whole genome duplication, and for a given region of maize, there will be a single purple line in sorghum.
All together, this means that there is a 2:4 syntenic relationshipt between sorghum and maize. There are two in sorghum form the pre-grass whole genome duplication event, and there are four in maize from the pre-grass whole genome duplication event combined with the subsequent maize-specific whole genome duplication event. This means that for any genomic region in maize or sorghum, there are 5 other syntenic regions. This gives rise for the possibility of comparing 6 syntenic regions at once: 2 from sorghum and 4 from maize.
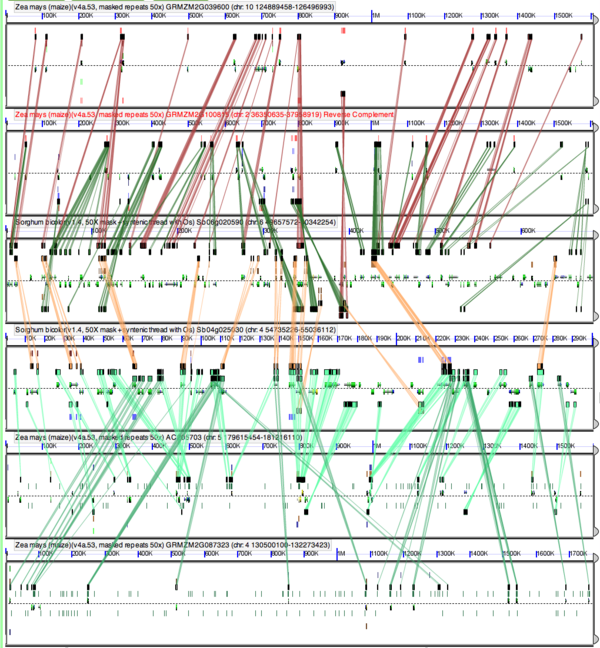
Another way to see these patterns is through high-resolution analysis of syntenic regions use GEvo. If SynMap is used to create and visualize syntenic dotplots, the results are interactive and provide links to GEvo. There is an example 6-way comparison of syntenic regions from maize sorghum dating back to the pre-grass whole genome duplication event. In this figure, the two sorghum regions are in the middle two regions, which two maize regions above or below each sorghum region. These maize regions are orthologous to the closest sorghum region, and the two sorghum regions are paralogous (or homeologous) to each other.
One additional feature of SynMap is that it will generate a summary table of all syntenic regions and the genes that are contained within each region. Each pair of genes will contain a link to GEvo. The file used in this example can be downloaded here: http://synteny.cnr.berkeley.edu/CoGe//diags/Sorghum_bicolor/Zea_mays_maize/6807_8082.CDS-CDS.blastn.dag_geneorder_D40_g20_A10.all.aligncoords.ks .