How to extract genomic features: Difference between revisions
Jump to navigation
Jump to search
| Line 7: | Line 7: | ||
===Find your genome of interest in [[OrganismView]]=== | ===Find your genome of interest in [[OrganismView]]=== | ||
[[Image:OrganismView Example Search.png|thumb|600px|right|Searching for the organism "Arabidopsis thaliana" in OrganismView]] | [[Image:OrganismView Example Search.png|thumb|600px|right|Searching for the organism "Arabidopsis thaliana" in OrganismView. Version 9 has been selected containing unmasked genomic sequence. Dataset chr1.xml and chromosome 1 have been selected.]] | ||
#Go to [[OrganismView ]] and search for your organism by name by typing part of its name in the name search box, or part of the [[organism description]] in the description box. | #Go to [[OrganismView ]] and search for your organism by name by typing part of its name in the name search box, or part of the [[organism description]] in the description box. | ||
#Select the correct organism from the organism list | #Select the correct organism from the organism list | ||
Revision as of 05:37, 2 January 2010
Overview
Extracting genomic features from a genomic region in a genome of interest is easy using GenomeView. All you need to do is:
- Find your genome of interest in OrganismView or your genomic feature in FeatView
- Visualize the genomic region of interest in GenomeView
- Select and export a genomic region from OrganismView for automatic feature extraction using FeatList
- Select the features of interest and send them to FastaView to get their sequences in fasta format
Find your genome of interest in OrganismView
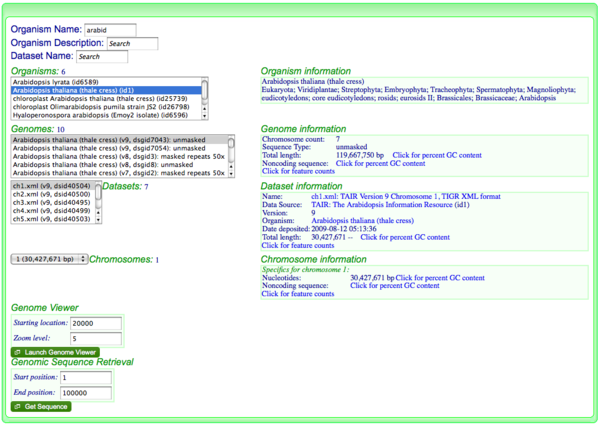
- Go to OrganismView and search for your organism by name by typing part of its name in the name search box, or part of the organism description in the description box.
- Select the correct organism from the organism list
- Select the appropriate genome from the list of genomes for that organism
- If needed, select the appropriate dataset and chromosome for the genome