FeatView: Difference between revisions
No edit summary |
|||
| Line 22: | Line 22: | ||
===Running a search=== | ===Running a search=== | ||
After the search criteria have been specified, just press the "Search". The results will come back and be displayed below the search criteria box. <font color=red>Warning:</font> this can take a while, especially if wildcards have been used. | After the search criteria have been specified, just press the "Search". The results will come back and be displayed below the search criteria box. <font color=red>Warning:</font> this can take a while, especially if wildcards have been used. | ||
=Annotation information and links for a selected genomic feature= | |||
==Buttons== | |||
===Get Seqeunce=== | |||
==In annotation links== | |||
==Why are there sometimes multiple annotations?== | |||
Multiple annotations are shown if there are multiple features of that type in that genome with the same name. All of these get displayed right after one another. A common case that causes this is when there are multiple splice variants of a gene, all of which share the same name | |||
Revision as of 06:14, 2 January 2010
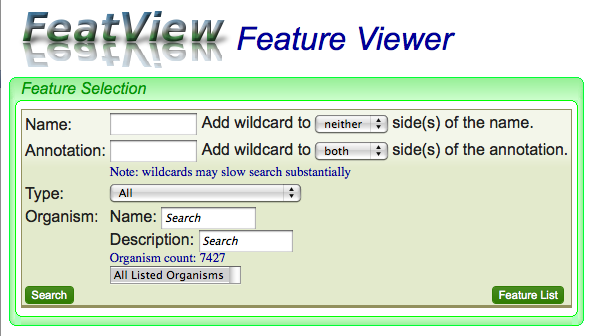
FeatView is CoGe's tool for finding genomic features by name or description.
WARNING: It is often difficult to find a genomic feature by name. The main reason for this is the variety of names used for any given genomic feature. While CoGe can store multiple names for a given genomic feature, only the names supplied in the original data file (dataset) are used. If you are having difficulty finding a genomic feature by name, try to find the sequence for the feature and use CoGeBlast to search that sequence against the genome to which it belongs.
Quick Start
To use, just:
- Type in the name of the genomic feature in the "Name:" text box.
- Click on the "Search" button.
- Select the appropriate feature type.
- Select the appropriate genome for the feature.
- The annotation for the feature appears at the bottom of the screen.
Searching
There are several options when searching for a genomic feature. The easiest and fastest is if you know the full name of the feature and don't use any wildcards.
Searching by name
Type in the name of the feature in the "Name:" text box
Searching by annotation
Using wildcards
Limiting search by feature type
Limiting search by organism
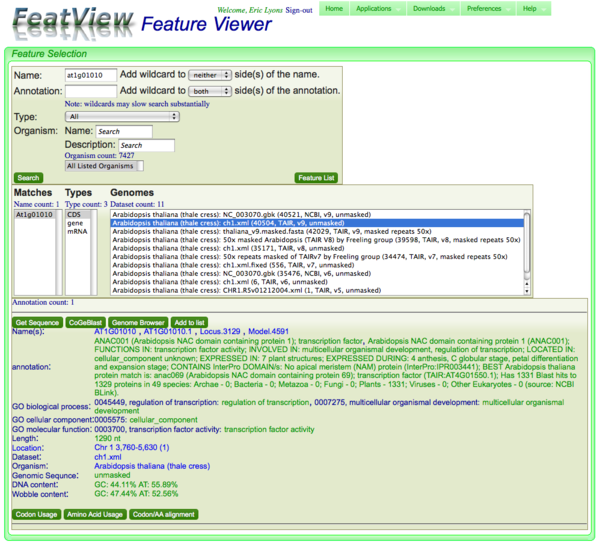
Running a search
After the search criteria have been specified, just press the "Search". The results will come back and be displayed below the search criteria box. Warning: this can take a while, especially if wildcards have been used.
Annotation information and links for a selected genomic feature
Buttons
Get Seqeunce
In annotation links
Why are there sometimes multiple annotations?
Multiple annotations are shown if there are multiple features of that type in that genome with the same name. All of these get displayed right after one another. A common case that causes this is when there are multiple splice variants of a gene, all of which share the same name