How to extract genomic features: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 10: | Line 10: | ||
#The genomic location by organism, chromosome, and nucleotide position | #The genomic location by organism, chromosome, and nucleotide position | ||
#The name of a [[genomic feature]] in the genomic region | #The name of a [[genomic feature]] in the genomic region | ||
<gallery widths=300px heights= | <gallery widths=300px heights=200px> | ||
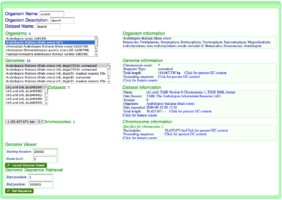
File:OrganismView Example Search.png|Searching for the organism "Arabidopsis thaliana" in OrganismView. Version 9 has been selected containing unmasked genomic sequence. Dataset ch1.xml and chromosome 1 have been selected. | File:OrganismView Example Search.png|Searching for the organism "Arabidopsis thaliana" in OrganismView. Version 9 has been selected containing unmasked genomic sequence. Dataset ch1.xml and chromosome 1 have been selected. | ||
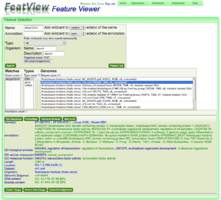
File:FeatView At1g01010.png|FeatView's results after searching for genomic features with the name "At1g01010". The Arabidopsis thaliana genome from TAIR version 9 is selected and the feature type "[[CDS]]" is selected. | File:FeatView At1g01010.png|FeatView's results after searching for genomic features with the name "At1g01010". The Arabidopsis thaliana genome from TAIR version 9 is selected and the feature type "[[CDS]]" is selected. | ||
Revision as of 18:03, 2 January 2010
Overview
Extracting genomic features from a genomic region in a genome of interest is easy using GenomeView. All you need to do is:
- Find your genome of interest in OrganismView or your genomic feature in FeatView
- Visualize the genomic region of interest in GenomeView
- Select and export a genomic region from OrganismView for automatic feature extraction using FeatList
- Select the features of interest and send them to FastaView to get their sequences in fasta format
Finding the genomic region of interest
There are two general ways to find a genomic region of interest depending on if you know:
- The genomic location by organism, chromosome, and nucleotide position
- The name of a genomic feature in the genomic region
-
Searching for the organism "Arabidopsis thaliana" in OrganismView. Version 9 has been selected containing unmasked genomic sequence. Dataset ch1.xml and chromosome 1 have been selected.
-
FeatView's results after searching for genomic features with the name "At1g01010". The Arabidopsis thaliana genome from TAIR version 9 is selected and the feature type "CDS" is selected.
Find your genome of interest in OrganismView
This option should be used if you know the organism, chromosome, and nucleotide position of the genomic region in which you are interested.
- Go to OrganismView and search for your organism by name by typing part of its name in the name search box, or part of the organism description in the description box. For more information about this, please see OrganismView.
- Select the correct organism from the organism list.
- Select the appropriate genome from the list of genomes for that organism.
- If needed, select the appropriate dataset and chromosome for the genome.
- Type in the appropriate location of the genomic region in which you are interested in the "Starting location" box under the "Genome Viewer" section in the lower right of the screen of OrganismView.
- Press the button called "Launch Genome Viewer" to launch GenomeView to display this genomic region.
Find your genomic feature in FeatView
This option should be used if you know the name of the genomic feature in which you are interested.
- Go to FeatView and search for your genomic feature by name or annotation. For more information about this, please see FeatView.
- Select the correct genome, dataset and genomic feature type for your genomic feature
- Press the "Genome Browser" button located above the displayed annotations for the selected genomic feature