Whole genome duplication: Difference between revisions
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
One common way to detect whole genome duplications is through [[syntenic dotplots]]. CoGe's tools [[SynMap]] provides an easy to use interface for generating [[syntenic dotplots]] between any two genomes stored in the [[CoGe database]]. [[SynMap#Example_Results | This example]], shows the results from [[SynMap]] to detect two ancient shared whole genome duplication events in the lineage of Arabidopsis. | One common way to detect whole genome duplications is through [[syntenic dotplots]]. CoGe's tools [[SynMap]] provides an easy to use interface for generating [[syntenic dotplots]] between any two genomes stored in the [[CoGe database]]. [[SynMap#Example_Results | This example]], shows the results from [[SynMap]] to detect two ancient shared whole genome duplication events in the lineage of Arabidopsis. | ||
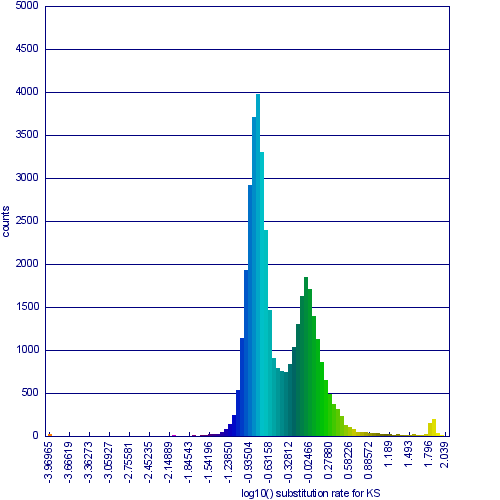
[[Image:Master 3068 8.CDS-CDS.blastn geneorder D20 g10 A5.w2000.ks.hist-1.png|thumb|center|800px|Figure 1b: Histogram of synonymous rate change data for syntenic gene pairs between Arabidopsis thaliana and Arabidopsis lyrata. The two obvious peaks in the distribution are from syntenic gene pairs [[(syntelogs)]] derived from the speciation of these two taxa and from their shared most recent whole genome duplication event, known as alpha.]] | |||
Revision as of 01:18, 11 January 2010
A whole genome duplication event is a genomic phenomena where an organism copies its entire genome, thereby creating a 2N ploidy where N was the ploidy prior to the duplication event. As many organisms that undergo these events are diploid, and hence go from 2N to 4N, whole genome duplication events are often referred to as tetraploidy events, although the nomenclature can be manipulated as per the number of haploid copies (i.e. hexaploidy for wheat).
One common way to detect whole genome duplications is through syntenic dotplots. CoGe's tools SynMap provides an easy to use interface for generating syntenic dotplots between any two genomes stored in the CoGe database. This example, shows the results from SynMap to detect two ancient shared whole genome duplication events in the lineage of Arabidopsis.