MaizeGDB and CoGe: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 13: | Line 13: | ||
===Automatically running [[GEvo]] with syntenic maize-sorghum regions pre-loaded=== | ===Automatically running [[GEvo]] with syntenic maize-sorghum regions pre-loaded=== | ||
[[Image:GEvo-running-maize-sorghum.png|thumb|800px|center|GEvo running a maize-sorghum syntenic analysis automatically. Genomic regions (400kb) have been preloaded in the sequence submission boxes. http://tinyurl.com/yhgnu8a]] | |||
===[[GEvo]]'s results=== | ===[[GEvo]]'s results=== | ||
Revision as of 20:11, 8 February 2010
Overview
MaizeGDB is linked to CoGe's through its genome browser. A link to CoGe will be shown at the top of the browser when viewing a genomic region from the maize genome assembly (i.e. pseudomolecules). This link will take you to the same region in CoGe's GenomeView. GenomeView will automatically find maize genes with annotations that link to GEvo for syntenic gene sets with sorghum, and show those genes with special glyphs. When you view the annotations for these glyphs (by clicking on them), there will be a link to GEvo. When clicked on, these GEvo links will launch GEvo with the syntenic maize-sorghum regions pre-loaded and automatically start a GEvo analysis using blastz.
MaizeGDB's genome browser link to CoGe's genome broswer, GenomeView

GenomeView showing glyphs for genes linked to GEvo
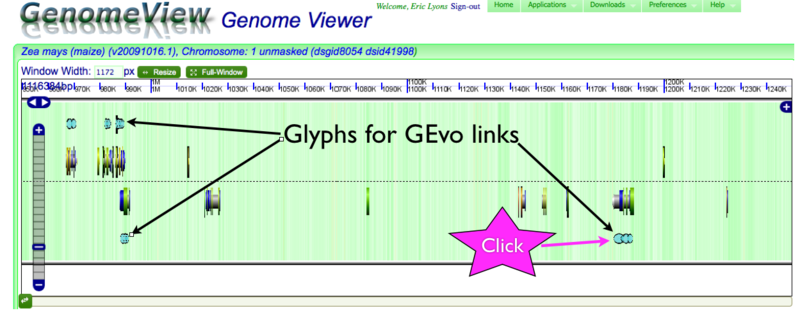
Getting a link to GEvo

Automatically running GEvo with syntenic maize-sorghum regions pre-loaded
