Flanking Gene Method: Difference between revisions
No edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
The flanking gene method is a procedure in which possible transposed genes are identified by comparing two neighboring [[syntenic]] gene pairs. An example visualization of this method is shown to the right. | The flanking gene method is a procedure in which possible transposed genes are identified by comparing two neighboring [[syntenic]] gene pairs. An example visualization of this method is shown to the right. | ||
Here, there are two pairs of syntenic genes (numbered 1 and 2). In the procedure, sequential, adjoining genes (genes 1 and 2) are compared between two species. If said genes are from orthologous regions, one would assume the genes remain sequential. However, in a transposition event another gene (eg, gene X in red) inserts itself between two otherwise sequential genes. In this case, gene X is a possible transposed gene, identified by its presence between genes 1 and 2 which would be otherwise adjoining each other. This method of identification is previously described in Freeling et al 2008. | Here, there are two pairs of syntenic genes (numbered gene 1 and gene 2). In the procedure, sequential, adjoining genes (genes 1 and 2) are compared between two species. If said genes are from orthologous regions, one would assume the genes remain sequential. However, in a transposition event another gene (eg, gene X in red) inserts itself between two otherwise sequential genes. In this case, gene X is a possible transposed gene, identified by its presence between genes 1 and 2 which would be otherwise adjoining each other. This method of identification is previously described in Freeling et al 2008. | ||
Further analysis uses additional an additional species as an outgroup to determine whether or not gene X is, in fact, an insertion between genes 1 an 2 rather than merely a gene in sequence that's been deleted. | Further analysis uses additional an additional species as an outgroup to determine whether or not gene X is, in fact, an insertion between genes 1 an 2 rather than merely a gene in sequence that's been deleted. | ||
Revision as of 19:49, 20 April 2010
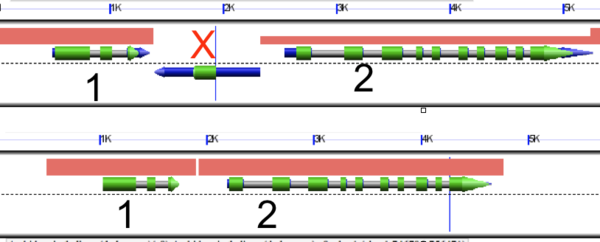
The flanking gene method is a procedure in which possible transposed genes are identified by comparing two neighboring syntenic gene pairs. An example visualization of this method is shown to the right.
Here, there are two pairs of syntenic genes (numbered gene 1 and gene 2). In the procedure, sequential, adjoining genes (genes 1 and 2) are compared between two species. If said genes are from orthologous regions, one would assume the genes remain sequential. However, in a transposition event another gene (eg, gene X in red) inserts itself between two otherwise sequential genes. In this case, gene X is a possible transposed gene, identified by its presence between genes 1 and 2 which would be otherwise adjoining each other. This method of identification is previously described in Freeling et al 2008.
Further analysis uses additional an additional species as an outgroup to determine whether or not gene X is, in fact, an insertion between genes 1 an 2 rather than merely a gene in sequence that's been deleted.