GC content shift: Difference between revisions
Jump to navigation
Jump to search
| Line 6: | Line 6: | ||
==Comparison of Schizosaccharomyces fungi japonicus strain yFS275 and pombe 972h-== | ==Comparison of Schizosaccharomyces fungi japonicus strain yFS275 and pombe 972h-== | ||
{| class="wikitable" style="text-align: center; width: 200px; height: 200px;" | |||
|+ Genome Overview | |||
|- | |||
! scope="col" | | |||
! scope="col" | japonicus | |||
! scope="col" | pombe | |||
|- | |||
! scope="row" | Length | |||
| 1 || 10,082,004 | |||
|- | |||
! scope="row" | GC% | |||
| 2 || 36 | |||
|- | |||
! scope="row" | AT% | |||
| 3 || 64 | |||
|- | |||
! scope="row" | CDS GC% | |||
| 4 || 39.5 | |||
|- | |||
! scope="row" | CDS AT% | |||
| 5 || 60.5 | |||
|- | |||
! scope="row" | Wobble GC% | |||
| 5 || 32.8 | |||
|- | |||
! scope="row" | Wobble AT% | |||
| 5 || 67.2 | |||
|} | |||
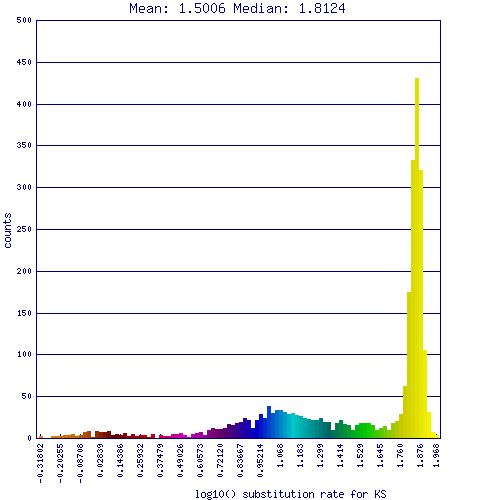
[[Image:Master 7763 1483.CDS-CDS.lastz.dag.go c4 D20 g10 A5.aligncoords.gcoords ct0.w1200.ks.png|thumb|right|500px|SynMap syntenic dotplot of two Schizosaccharomyces fungi: japonicus strain yFS275 (x-axis) and pombe 972h (y-axis). Syntenic gene pairs are colored based on their synonymous mutation rates (see histogram below). While there have been many genomic rearrangements since these lineages diverged, nearly every genomic regions in both species have a syntenic partner region (albeit it small regions of contiguous synteny). Results may be regenerated at: http://genomevolution.org/r/pi4]] | [[Image:Master 7763 1483.CDS-CDS.lastz.dag.go c4 D20 g10 A5.aligncoords.gcoords ct0.w1200.ks.png|thumb|right|500px|SynMap syntenic dotplot of two Schizosaccharomyces fungi: japonicus strain yFS275 (x-axis) and pombe 972h (y-axis). Syntenic gene pairs are colored based on their synonymous mutation rates (see histogram below). While there have been many genomic rearrangements since these lineages diverged, nearly every genomic regions in both species have a syntenic partner region (albeit it small regions of contiguous synteny). Results may be regenerated at: http://genomevolution.org/r/pi4]] | ||
Revision as of 22:56, 25 August 2010
GC content shift is an evolutionary genome-wide change in the GC content of an organism. The evolutionary consequences of such a change is unknown. They are detected by comparing the genomes of two related organisms that share a recent common ancestor, and determining if their have different:
- AT/GC genomic composition
- High rate of synonymous mutations for syntenic gene pairs
- Amino acid usage frequencies
- Codon usage frequencies
Comparison of Schizosaccharomyces fungi japonicus strain yFS275 and pombe 972h-
| japonicus | pombe | |
|---|---|---|
| Length | 1 | 10,082,004 |
| GC% | 2 | 36 |
| AT% | 3 | 64 |
| CDS GC% | 4 | 39.5 |
| CDS AT% | 5 | 60.5 |
| Wobble GC% | 5 | 32.8 |
| Wobble AT% | 5 | 67.2 |