GC content shift: Difference between revisions
Jump to navigation
Jump to search
| Line 38: | Line 38: | ||
|- | |- | ||
! scope="row" | Codon Usage Table | ! scope="row" | Codon Usage Table | ||
| [[Image:Schizosaccharomyces japonicus strain yFS275 amino acid usage table.png|thumb|center| | | [[Image:Schizosaccharomyces japonicus strain yFS275 amino acid usage table.png|thumb|center|100px]] || [[Image:Schizosaccharomyces pombe 972h amino acid usage table.png|thumb|center|100px]] | ||
|} | |} | ||
Revision as of 00:13, 26 August 2010
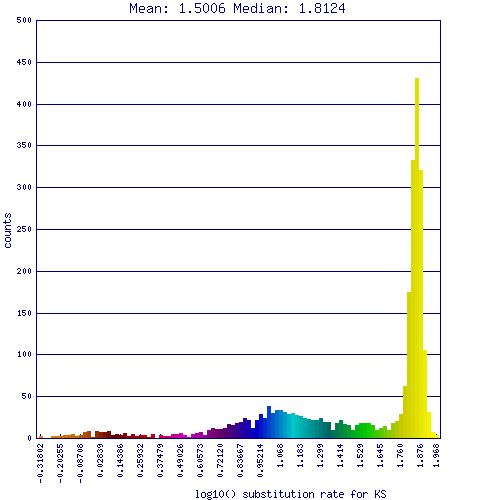
GC content shift is an evolutionary genome-wide change in the GC content of an organism. The evolutionary consequences of such a change is unknown. They are detected by comparing the genomes of two related organisms that share a recent common ancestor, and determining if their have different:
- AT/GC genomic composition
- High rate of synonymous mutations for syntenic gene pairs
- Amino acid usage frequencies
- Codon usage frequencies
Comparison of Schizosaccharomyces fungi japonicus strain yFS275 and pombe 972h-
| japonicus | pombe | |
|---|---|---|
| Length | 11,300,431 | 10,082,004 |
| GC% | 42.6 | 36 |
| AT% | 54.9 | 64 |
| CDS GC% | 45.5 | 39.5 |
| CDS AT% | 54.5 | 60.5 |
| Wobble GC% | 44.1 | 32.8 |
| Wobble AT% | 55.9 | 67.2 |
| Codon Usage Table | 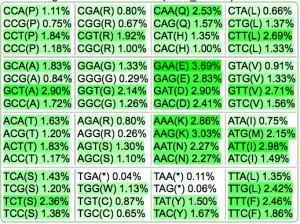 |
 |
| Codon Usage Table |  |
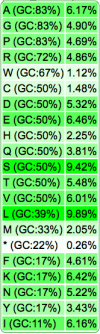 |