GEvo Blastn Bug: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
|||
| Line 1: | Line 1: | ||
==Bug Description== | == Bug Description == | ||
==Visualization== | When using blastn in [[GEvo]], a large HSP appear in the middle of the region when the query sequence length is changed by 1 nucleotide. | ||
Difference between analyses is that addition of 1 nucleotide to the top panel of the top analysis | |||
[[ | == Visualization == | ||
[[ | |||
Difference between analyses is that addition of 1 nucleotide to the top panel of the top analysis [[Image:Screen Shot 2012-02-11 at 4.55.25 AM.png|thumb|center|500px]] [[Image:Screen Shot 2012-02-11 at 4.57.06 AM.png|thumb|center|500px]] | |||
== Report of "disappearing" HSP in blast file == | |||
Top portion of blast report which contains the large HSP:<br> | |||
<pre>BLASTN 2.2.24+ | |||
Query= AT1G75520 | |||
Length=478 | |||
Subject= Bra008203 | |||
Length=17361 | |||
Score = 68.0 bits (35), Expect = 3e-14 | |||
Identities = 76/94 (80%), Gaps = 4/94 (4%) | |||
Strand=Plus/Plus | |||
Query 1 CTAGGTTTCGTGTTCCACTGATCAAAGATTTGAAAAAAAACATATACTTAGTAAACTTCA 60 | |||
|||||||| ||||||||||||||||||| || || ||||||| | |||||| | | |||
Sbjct 9943 CTAGGTTTTGTGTTCCACTGATCAAAGAGTT----AAGAACATATTTCAATAAAACTTTA 9998 | |||
Query 61 AGCAATTTTTATATTACCCAATTGAATTTCTCCA 94 | |||
| |||||||||| |||||| ||||||||||||| | |||
Sbjct 9999 AATAATTTTTATACTACCCAGTTGAATTTCTCCA 10032 | |||
Score = 54.5 bits (28), Expect = 3e-10 | |||
Identities = 32/34 (94%), Gaps = 0/34 (0%) | |||
Strand=Plus/Plus | |||
Query 442 TTGGCGTAGATGAATGTAAACGGATGGTAATATA 475 | |||
|||||||| |||||||||||||||| |||||||| | |||
Sbjct 10342 TTGGCGTATATGAATGTAAACGGATAGTAATATA 10375 | |||
Score = 50.7 bits (26), Expect = 4e-09 | |||
Identities = 77/95 (81%), Gaps = 5/95 (5%) | |||
Strand=Plus/Plus | |||
Query 129 ATATATATATACCCAACAACTGAGAAAAGATGGAAAAAGTTTAGTTAAAAACTGGTCCTG 188 | |||
||||||||||| |||||| || |||||||||||| || || ||||||||| | | | |||
Sbjct 10072 ATATATATATAACCAACATCTCGGAAAAGATGGAACAAATT-AGTTAAAAAAA---CATT 10127 | |||
Query 189 GGCGGCTTTAAATTATATTTATGCACTTAAATTTA 223 | |||
||||||||||||| ||||| ||| | ||||||||| | |||
Sbjct 10128 GGCGGCTTTAAATCATATTCATGTA-TTAAATTTA 10161 | |||
Score = 48.8 bits (25), Expect = 2e-08 | |||
Identities = 47/58 (81%), Gaps = 0/58 (0%) | |||
Strand=Plus/Plus | |||
Query 316 GGTTTTTACTTAGATAATATCGTGTCATTCCATCTAGATTCAACCCCTGTCTACAATA 373 | |||
|||||| ||| |||| |||| || || | | ||||||||||||||| ||||||||| | |||
Sbjct 10207 GGTTTTCACTGAGATGATATTGTTCCAGTTCCACTAGATTCAACCCCTCTCTACAATA 10264 | |||
Score = 25.7 bits (13), Expect = 0.15 | |||
Identities = 13/13 (100%), Gaps = 0/13 (0%) | |||
Strand=Plus/Plus | |||
Query 127 TAATATATATATA 139 | |||
||||||||||||| | |||
Sbjct 7327 TAATATATATATA 7339 | |||
</pre> | |||
Revision as of 12:00, 11 February 2012
Bug Description
When using blastn in GEvo, a large HSP appear in the middle of the region when the query sequence length is changed by 1 nucleotide.
Visualization
Difference between analyses is that addition of 1 nucleotide to the top panel of the top analysis

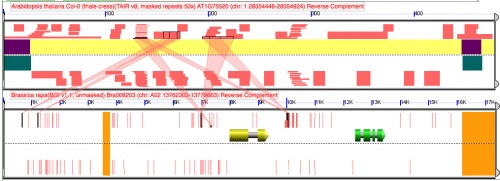
Report of "disappearing" HSP in blast file
Top portion of blast report which contains the large HSP:
BLASTN 2.2.24+
Query= AT1G75520
Length=478
Subject= Bra008203
Length=17361
Score = 68.0 bits (35), Expect = 3e-14
Identities = 76/94 (80%), Gaps = 4/94 (4%)
Strand=Plus/Plus
Query 1 CTAGGTTTCGTGTTCCACTGATCAAAGATTTGAAAAAAAACATATACTTAGTAAACTTCA 60
|||||||| ||||||||||||||||||| || || ||||||| | |||||| |
Sbjct 9943 CTAGGTTTTGTGTTCCACTGATCAAAGAGTT----AAGAACATATTTCAATAAAACTTTA 9998
Query 61 AGCAATTTTTATATTACCCAATTGAATTTCTCCA 94
| |||||||||| |||||| |||||||||||||
Sbjct 9999 AATAATTTTTATACTACCCAGTTGAATTTCTCCA 10032
Score = 54.5 bits (28), Expect = 3e-10
Identities = 32/34 (94%), Gaps = 0/34 (0%)
Strand=Plus/Plus
Query 442 TTGGCGTAGATGAATGTAAACGGATGGTAATATA 475
|||||||| |||||||||||||||| ||||||||
Sbjct 10342 TTGGCGTATATGAATGTAAACGGATAGTAATATA 10375
Score = 50.7 bits (26), Expect = 4e-09
Identities = 77/95 (81%), Gaps = 5/95 (5%)
Strand=Plus/Plus
Query 129 ATATATATATACCCAACAACTGAGAAAAGATGGAAAAAGTTTAGTTAAAAACTGGTCCTG 188
||||||||||| |||||| || |||||||||||| || || ||||||||| | |
Sbjct 10072 ATATATATATAACCAACATCTCGGAAAAGATGGAACAAATT-AGTTAAAAAAA---CATT 10127
Query 189 GGCGGCTTTAAATTATATTTATGCACTTAAATTTA 223
||||||||||||| ||||| ||| | |||||||||
Sbjct 10128 GGCGGCTTTAAATCATATTCATGTA-TTAAATTTA 10161
Score = 48.8 bits (25), Expect = 2e-08
Identities = 47/58 (81%), Gaps = 0/58 (0%)
Strand=Plus/Plus
Query 316 GGTTTTTACTTAGATAATATCGTGTCATTCCATCTAGATTCAACCCCTGTCTACAATA 373
|||||| ||| |||| |||| || || | | ||||||||||||||| |||||||||
Sbjct 10207 GGTTTTCACTGAGATGATATTGTTCCAGTTCCACTAGATTCAACCCCTCTCTACAATA 10264
Score = 25.7 bits (13), Expect = 0.15
Identities = 13/13 (100%), Gaps = 0/13 (0%)
Strand=Plus/Plus
Query 127 TAATATATATATA 139
|||||||||||||
Sbjct 7327 TAATATATATATA 7339