Nelumbo v. Vitis: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
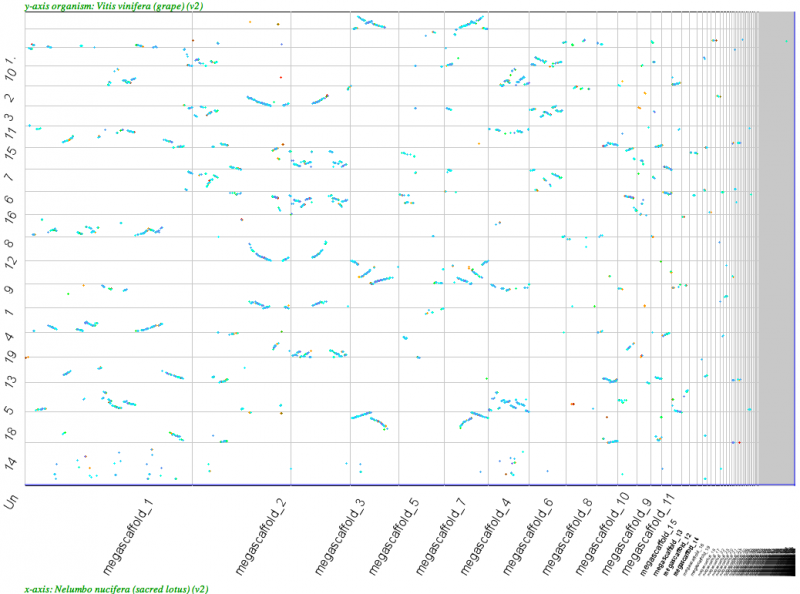
[[File:Screen Shot 2012-03-21 at 1.15.17 PM.png|thumb|center|800px|[[Syntenic dotplot]] of Nelumbo megascaffolds (x-axis) v. Vitus (y-axis). Note the 2xNelumbo to 3xVitus syntenic relationship. Results may be regenerated: http://genomevolution.org/r/4mrm]] | [[File:Screen Shot 2012-03-21 at 1.15.17 PM.png|thumb|center|800px|[[Syntenic dotplot]] of Nelumbo megascaffolds (x-axis) v. Vitus (y-axis). Note the 2xNelumbo to 3xVitus syntenic relationship. Results may be regenerated: http://genomevolution.org/r/4mrm]] | ||
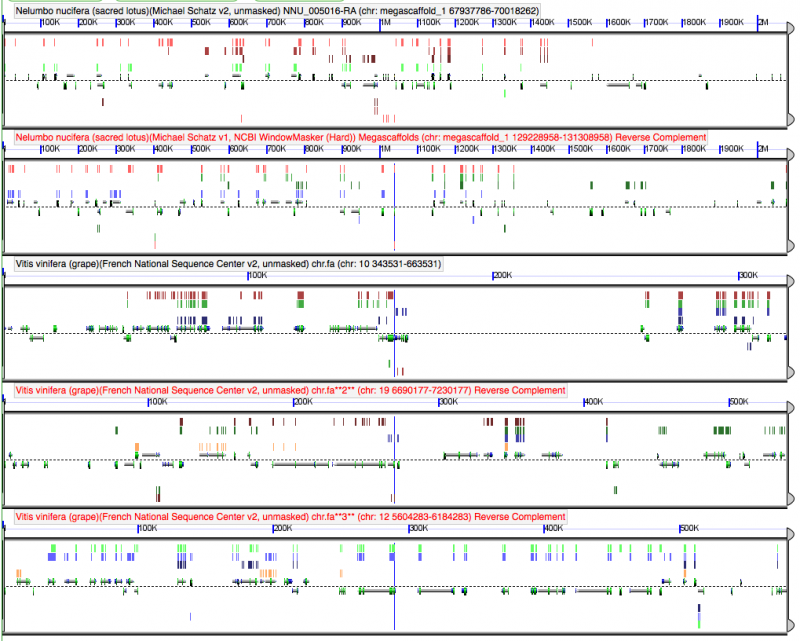
[[File:Screen shot 2012-04-09 at 6.00.09 PM.png|thumb|center|800px[[GEvo]] analysis of Nelumbo (2x; top two panels) v. Vitis (3x; bottom three panels). All regions show [[fractionation]] with respect to one another. Evidence that [[tetraploidy]] in Nelumbo and [[hexaploidy]] in Vitis are independently derived.]] | [[File:Screen shot 2012-04-09 at 6.00.09 PM.png|thumb|center|800px|[[GEvo]] analysis of Nelumbo (2x; top two panels) v. Vitis (3x; bottom three panels). All regions show [[fractionation]] with respect to one another. Evidence that [[tetraploidy]] in Nelumbo and [[hexaploidy]] in Vitis are independently derived.]] | ||
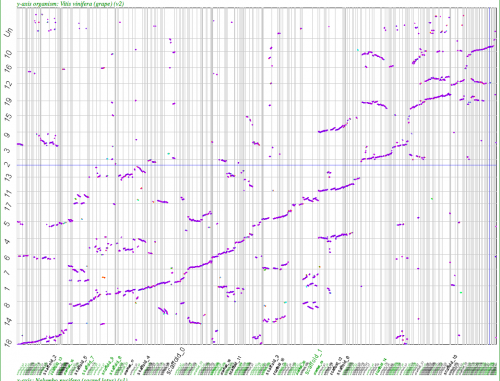
[[File:Screen shot 2012-02-27 at 2.01.45 PM.png|thumb|center|800px|Syntenic dotplot of Nelumbo v. Vitis. Note the 2xNelumbo to 3xVitis syntenic relationship. Syntenic gene pairs colored by Ks values (all are approximately the same). Evidence that duplications are independent of one another. Results may be regenerated: http://genomevolution.org/r/4kei]] | [[File:Screen shot 2012-02-27 at 2.01.45 PM.png|thumb|center|800px|Syntenic dotplot of Nelumbo v. Vitis. Note the 2xNelumbo to 3xVitis syntenic relationship. Syntenic gene pairs colored by Ks values (all are approximately the same). Evidence that duplications are independent of one another. Results may be regenerated: http://genomevolution.org/r/4kei]] | ||
Revision as of 01:02, 10 April 2012