Multidimensional whole genome synteny: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 4: | Line 4: | ||
|- | |- | ||
| | | | ||
| [File:Screen Shot 2013-09-04 at 10.35.32 AM.png|thumb|400px]] | | [[File:Screen Shot 2013-09-04 at 10.35.32 AM.png|thumb|400px]] | ||
|- | |- | ||
| | | | ||
Revision as of 17:39, 4 September 2013
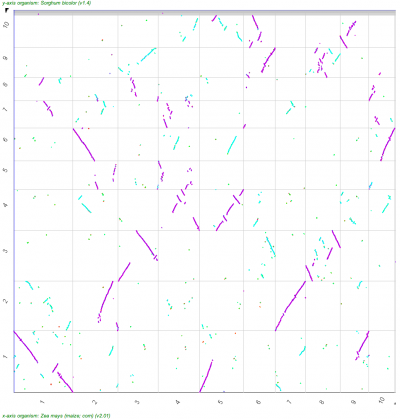
A popular means to compare whole genomes is the use of Syntenic dotplots. These dotplots reveal regions of the genomes that are related through common ancestry -- either through the divergence of lineages/species or through intra-genomic duplications. CoGe makes the generation of these available through its tool SynMap as well as providing the means to add additional information about the divergence age of syntenic gene pairs to the dots in the dotplot.