Install coge: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
== Server with existing CoGe Installation == | == Server with existing CoGe Installation == | ||
=== Create new mysql database === | === Create new mysql database === | ||
Note: be sure to disable apparmour for MySQL. | Note: be sure to disable apparmour for MySQL. | ||
Dump CoGe Database schema | Dump CoGe Database schema | ||
* File download: http://data.iplantcollaborative.org/quickshare/71b18508287f9fb0/cogetable.sql (AUTO_INCREMENT removed) | |||
*File download: http://data.iplantcollaborative.org/quickshare/71b18508287f9fb0/cogetable.sql (AUTO_INCREMENT removed) | |||
mysqldump -d -h localhost -u root -pXXXXXXX coge | sed 's/AUTO_INCREMENT=[0-9]*\b/AUTO_INCREMENT/' > cogetable.sql | mysqldump -d -h localhost -u root -pXXXXXXX coge | sed 's/AUTO_INCREMENT=[0-9]*\b/AUTO_INCREMENT/' > cogetable.sql | ||
Create new CoGe Database | Create new CoGe Database | ||
create database oryza_coge | create database oryza_coge | ||
Initialize new coge database | Initialize new coge database | ||
mysql -u root -pXXXXXXXX oryza_coge < cogetable.sql | mysql -u root -pXXXXXXXX oryza_coge < cogetable.sql | ||
Populate a few entries in the feature_type table | Populate a few entries in the feature_type table | ||
* This is important because part of CoGe's code-base is keyed to feature_type_ids. This is done in order to improve performance of the system by using a feature_type_id to retrieve features of a particular type. An example is OrganismView which needs to find features of type "chromosome" in order to determine the size of a genome. The table loaded here contains [[CoGe's 10 base feature types|10 feature types]] | |||
*This is important because part of CoGe's code-base is keyed to feature_type_ids. This is done in order to improve performance of the system by using a feature_type_id to retrieve features of a particular type. An example is OrganismView which needs to find features of type "chromosome" in order to determine the size of a genome. The table loaded here contains [[CoGe's 10 base feature types|10 feature types]] | |||
*Download file: http://data.iplantcollaborative.org/quickshare/ac8758f83c9b29b1/feature_type.sql | *Download file: http://data.iplantcollaborative.org/quickshare/ac8758f83c9b29b1/feature_type.sql | ||
| Line 25: | Line 27: | ||
Create new user for new CoGe database | Create new user for new CoGe database | ||
*Want a web-user with limited write privileges and a power user to load new data | *Want a web-user with limited write privileges and a power user to load new data | ||
[[ | |||
[[Image:Screen shot 2012-07-09 at 11.24.25 AM.png|thumb|right|400px]] | |||
create user 'oryza_coge'@'localhost' IDENTIFIED BY 'XXXXXX'; | create user 'oryza_coge'@'localhost' IDENTIFIED BY 'XXXXXX'; | ||
| Line 34: | Line 38: | ||
flush privileges; | flush privileges; | ||
Note: | Note: The CoGe web-user needs edit/insert permission on some tables. Here is a snapshot of what these are: | ||
=== Deploy new CoGe Web-site === | === Deploy new CoGe Web-site === | ||
Download CoGe repository from GitHub | Download CoGe repository from GitHub | ||
git clone https://github.com/LyonsLab/coge.git | git clone https://github.com/LyonsLab/coge.git | ||
Make directories where files go and give write persmission | Make directories where files go and give write persmission | ||
cd coge/web | cd coge/web | ||
./setup.sh | ./setup.sh | ||
Configure apache | Configure apache | ||
sudo vi /etc/apache2/sites-enabled/000-default | sudo vi /etc/apache2/sites-enabled/000-default | ||
| Line 57: | Line 61: | ||
Alias /ocoge "/opt/apache2/Oryza_CoGe" | Alias /ocoge "/opt/apache2/Oryza_CoGe" | ||
<Directory "/opt/apache2/Oryza_CoGe"> | <Directory "/opt/apache2/Oryza_CoGe"> | ||
PerlInitHandler Apache2::Reload | |||
Options Includes Indexes ExecCGI FollowSymLinks | |||
AllowOverride AuthConfig | |||
SetEnv HOME "/opt/apache2/Oryza_CoGe/" | |||
Order allow,deny | |||
Allow from all | |||
</Directory> | </Directory> | ||
<Directory /home/mbomhoff/public/coge/services/JBrowse/JBrowse_TrackContent_WS/> | |||
Options FollowSymLinks +ExecCGI | |||
AddHandler wsgi-script .py | |||
</Directory> | |||
Alias /coge/services/JBrowse/track /home/mbomhoff/public/coge/services/JBrowse/JBrowse_TrackContent_WS/source.py | Alias /coge/services/JBrowse/track /home/mbomhoff/public/coge/services/JBrowse/JBrowse_TrackContent_WS/source.py | ||
<Directory /opt/apache2/Oryza_CoGe/services/JBrowse/JBrowse_TrackContent_WS/> | |||
Options FollowSymLinks +ExecCGI | |||
AddHandler wsgi-script .py | |||
</Directory> | |||
Alias /Oryza_CoGe/services/JBrowse/track /Oryza_CoGe/services/JBrowse/JBrowse_TrackContent_WS/source.py | Alias /Oryza_CoGe/services/JBrowse/track /Oryza_CoGe/services/JBrowse/JBrowse_TrackContent_WS/source.py | ||
##configure gobe (visualization tool for GEvo) | ##configure gobe (visualization tool for GEvo) | ||
<Directory "/opt/apache2/Oryza_CoGe/gobe/"> | |||
Options FollowSymLinks +ExecCGI | |||
AddHandler wsgi-script .py | |||
</Directory> | |||
Configure coge.conf file | Configure coge.conf file | ||
##This is a configuration file for CoGe. | ##This is a configuration file for CoGe. | ||
| Line 138: | Line 142: | ||
IMAGE_CACHE /opt/apache/Oryza_CoGe/data/image_cache/ | IMAGE_CACHE /opt/apache/Oryza_CoGe/data/image_cache/ | ||
===Install Perl Modules=== | === Install Perl Modules === | ||
*Install CoGe modules /modules directory of CoGe install path | *Install CoGe modules /modules directory of CoGe install path | ||
perl Makefile.PL lib=/usr/local/lib/perl; make install | perl Makefile.PL lib=/usr/local/lib/perl; make install | ||
*Install non-standard Perl modules | *Install non-standard Perl modules | ||
sudo apt-get install expat libexpat1-dev | DBIx::Class | ||
AuthCAS | |||
CGI::Log | |||
DBIx::Class::Schema | |||
Data::UUID | |||
Data::GUID | |||
sudo apt-get install expat libexpat1-dev | |||
XML::SAX::Expat | XML::SAX::Expat | ||
XML::Parser | |||
XML::Simple | |||
=== Install JBrowse === | IPC::System::Simple | ||
=== Install JBrowse === | |||
Download and install the JBrowse package from http://jbrowse.org/install/ | Download and install the JBrowse package from http://jbrowse.org/install/ | ||
| Line 166: | Line 180: | ||
mv JBrowse-1.9.7 jbrowse | mv JBrowse-1.9.7 jbrowse | ||
===Populate with test data=== | === Populate with test data === | ||
scripts/replicate_genome_between_coge_installations.pl -dsgid 11022 -u1 coge -p1 XXXXXXX -db1 coge -u2 oryza_coge -p2 XXXXXXX -db2 oryza_coge -sd /opt/apache/oryz_coge/data/genomic_sequence | scripts/replicate_genome_between_coge_installations.pl -dsgid 11022 -u1 coge -p1 XXXXXXX -db1 coge -u2 oryza_coge -p2 XXXXXXX -db2 oryza_coge -sd /opt/apache/oryz_coge/data/genomic_sequence | ||
==Troubleshooting== | == Troubleshooting == | ||
===Visualization in [[GEvo]] does not work=== | |||
This relies on a system known as [[Gobe]]. | === Visualization in [[GEvo]] does not work === | ||
* Apache configuration for gobe | |||
* Check to see if paths hard-coded into gobe/flash/service.wsgi need to be updated | This relies on a system known as [[Gobe]]. Check the following things: | ||
** NOTE: Not sure if this is required | |||
*Apache configuration for gobe | |||
*Check to see if paths hard-coded into gobe/flash/service.wsgi need to be updated | |||
**NOTE: Not sure if this is required | |||
Revision as of 17:53, 28 October 2013
Server with existing CoGe Installation
Create new mysql database
Note: be sure to disable apparmour for MySQL.
Dump CoGe Database schema
- File download: http://data.iplantcollaborative.org/quickshare/71b18508287f9fb0/cogetable.sql (AUTO_INCREMENT removed)
mysqldump -d -h localhost -u root -pXXXXXXX coge | sed 's/AUTO_INCREMENT=[0-9]*\b/AUTO_INCREMENT/' > cogetable.sql
Create new CoGe Database
create database oryza_coge
Initialize new coge database
mysql -u root -pXXXXXXXX oryza_coge < cogetable.sql
Populate a few entries in the feature_type table
- This is important because part of CoGe's code-base is keyed to feature_type_ids. This is done in order to improve performance of the system by using a feature_type_id to retrieve features of a particular type. An example is OrganismView which needs to find features of type "chromosome" in order to determine the size of a genome. The table loaded here contains 10 feature types
- Download file: http://data.iplantcollaborative.org/quickshare/ac8758f83c9b29b1/feature_type.sql
mysql -u root -pXXXXXXXXX oryza_coge < feature_type.sql
Create new user for new CoGe database
- Want a web-user with limited write privileges and a power user to load new data
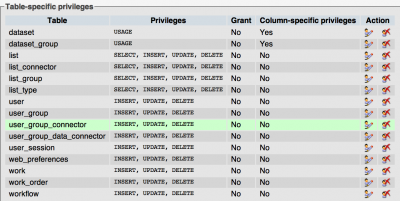
create user 'oryza_coge'@'localhost' IDENTIFIED BY 'XXXXXX'; grant all privileges on oryza_coge.* to oryza_coge; create user 'oryza_coge_web'@'localhost' IDENTIFIED BY 'XXXXXX'; grant select on oryza_coge.* to oryza_coge_web; flush privileges;
Note: The CoGe web-user needs edit/insert permission on some tables. Here is a snapshot of what these are:
Deploy new CoGe Web-site
Download CoGe repository from GitHub
git clone https://github.com/LyonsLab/coge.git
Make directories where files go and give write persmission
cd coge/web ./setup.sh
Configure apache
sudo vi /etc/apache2/sites-enabled/000-default
##Configure the main website Alias /Oryza_CoGe "/opt/apache2/Oryza_CoGe" Alias /oryza_coge "/opt/apache2/Oryza_CoGe" Alias /oCoGe "/opt/apache2/Oryza_CoGe" Alias /ocoge "/opt/apache2/Oryza_CoGe" <Directory "/opt/apache2/Oryza_CoGe"> PerlInitHandler Apache2::Reload Options Includes Indexes ExecCGI FollowSymLinks AllowOverride AuthConfig SetEnv HOME "/opt/apache2/Oryza_CoGe/" Order allow,deny Allow from all </Directory> <Directory /home/mbomhoff/public/coge/services/JBrowse/JBrowse_TrackContent_WS/> Options FollowSymLinks +ExecCGI AddHandler wsgi-script .py </Directory> Alias /coge/services/JBrowse/track /home/mbomhoff/public/coge/services/JBrowse/JBrowse_TrackContent_WS/source.py <Directory /opt/apache2/Oryza_CoGe/services/JBrowse/JBrowse_TrackContent_WS/> Options FollowSymLinks +ExecCGI AddHandler wsgi-script .py </Directory> Alias /Oryza_CoGe/services/JBrowse/track /Oryza_CoGe/services/JBrowse/JBrowse_TrackContent_WS/source.py ##configure gobe (visualization tool for GEvo) <Directory "/opt/apache2/Oryza_CoGe/gobe/"> Options FollowSymLinks +ExecCGI AddHandler wsgi-script .py </Directory>
Configure coge.conf file
##This is a configuration file for CoGe. ##Key Value pairs: ##<NAME> <PATH> #database configuration DBNAME oryza_coge DBHOST localhost DBPORT 3306 DBUSER oryza_coge_web DBPASS XXXXXXXXXXXX #basedir for coge COGEDIR /opt/apache/Oryza_CoGe/ #bin dir for coge's programs BINDIR /opt/apache/Oryza_CoGe/bin/ #data dir for coge's programs DATADIR /opt/apache/Oryza_CoGe/data/ #dir for pair-wise whole genome comparisons (e.g. SynMap) DIAGSDIR /opt/apache/Oryza_CoGe/diags/ #fasta dir FASTADIR /opt/apache/Oryza_CoGe/data/fasta/ #TMPL dir for coge's web page templates TMPLDIR /opt/apache/Oryza_CoGe/tmpl/ #temp dir for coge TEMPDIR /opt/apache/Oryza_CoGe/tmp/ #Base URL for web-site URL /Oryza_CoGe/ #URL for temp directory TEMPURL /Oryza_CoGe/tmp/ #blast style scoring matrix dirs BLASTMATRIX /opt/apache/Oryza_CoGe/data/blast/matrix/ #blastable DB BLASTDB /opt/apache/Oryza_CoGe/data/blast/db/ #directory for bed files BEDDIR /opt/apache/Oryza_CoGe/data/bed/ #servername for links SERVER http://coge.iplantcollaborative.org/Oryza_CoGe/ #directory for caching genome browser images IMAGE_CACHE /opt/apache/Oryza_CoGe/data/image_cache/
Install Perl Modules
- Install CoGe modules /modules directory of CoGe install path
perl Makefile.PL lib=/usr/local/lib/perl; make install
- Install non-standard Perl modules
DBIx::Class
AuthCAS
CGI::Log
DBIx::Class::Schema
Data::UUID
Data::GUID
sudo apt-get install expat libexpat1-dev
XML::SAX::Expat
XML::Parser
XML::Simple
IPC::System::Simple
Install JBrowse
Download and install the JBrowse package from http://jbrowse.org/install/
cd js unzip JBrowse-1.9.7.zip mv JBrowse-1.9.7 jbrowse
Populate with test data
scripts/replicate_genome_between_coge_installations.pl -dsgid 11022 -u1 coge -p1 XXXXXXX -db1 coge -u2 oryza_coge -p2 XXXXXXX -db2 oryza_coge -sd /opt/apache/oryz_coge/data/genomic_sequence
Troubleshooting
Visualization in GEvo does not work
This relies on a system known as Gobe. Check the following things:
- Apache configuration for gobe
- Check to see if paths hard-coded into gobe/flash/service.wsgi need to be updated
- NOTE: Not sure if this is required