GenomeView: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
GenomeView is the EPIC-CoGe genome browser that allows users to navigate genome sequence, annotation, and associated data tracks (quantitative measurements, SNPs, alignments). | GenomeView is the EPIC-CoGe genome browser that allows users to navigate genome sequence, annotation, and associated data tracks (quantitative measurements, SNPs, alignments). | ||
<pre style="color:red"> | |||
Note: this article is still under development. | |||
If you have any questions or comments please contact us directly at coge.genome@gmail.com | |||
</pre> | |||
More information, including video tutorials, can be found at [[EPIC-CoGe]]. | |||
= Navigation = | = Navigation = | ||
Revision as of 19:15, 16 January 2015
Overview
GenomeView is the EPIC-CoGe genome browser that allows users to navigate genome sequence, annotation, and associated data tracks (quantitative measurements, SNPs, alignments).
Note: this article is still under development. If you have any questions or comments please contact us directly at coge.genome@gmail.com
More information, including video tutorials, can be found at EPIC-CoGe.
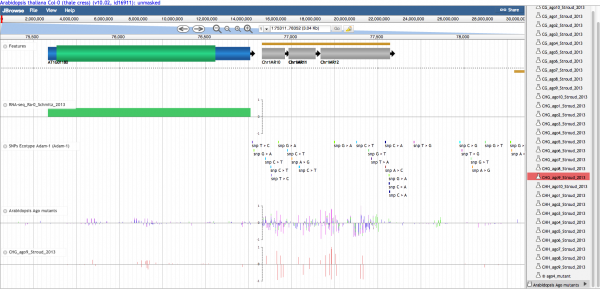
Panning left-right
There are several ways to move along a genomic region:
- Click on the track and drag with mouse
- Use the left/right arrows located on the left of the viewer
- Click a location in the macro or micro coordinate bars at the top of the viewer.
- Enter a chromosome/start/end location in the location input bar at the top of the viewer.
Zooming in and out
You can zoom into and out of a genomic region by:
- Click the (+) and (-) magnifying glass icons at the top of the viewer.
- Double-clicking on a region will zoom in one level
Selecting Tracks
Use the track selector menu on the right of the viewer to add or remove tracks to the current view.