GenomeView: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
|||
| Line 1: | Line 1: | ||
= Overview = | = Overview = | ||
GenomeView is the EPIC-CoGe genome browser that allows users to navigate genome sequence, annotation, and associated data tracks (quantitative measurements, SNPs, alignments). | GenomeView is also known as the [[EPIC-CoGe]] genome browser that allows users to navigate genome sequence, annotation, and associated data tracks (quantitative measurements, SNPs, alignments, and markers). | ||
More information, including video tutorials, can be found at [[EPIC-CoGe]]. | More information, including video tutorials, can be found at [[EPIC-CoGe]]. | ||
Revision as of 17:08, 15 September 2016
Overview
GenomeView is also known as the EPIC-CoGe genome browser that allows users to navigate genome sequence, annotation, and associated data tracks (quantitative measurements, SNPs, alignments, and markers).
More information, including video tutorials, can be found at EPIC-CoGe.
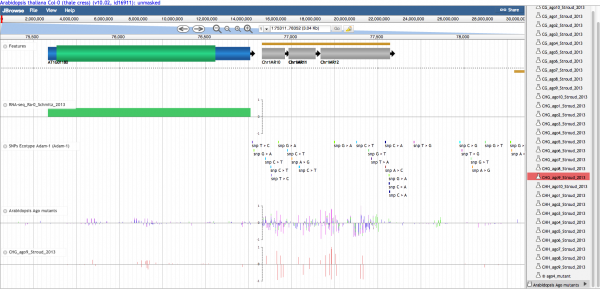
Panning left-right
There are several ways to move along a genomic region:
- Click on the track and drag with mouse
- Use the left/right arrows located on the left of the viewer
- Click a location in the macro or micro coordinate bars at the top of the viewer.
- Enter a chromosome/start/end location in the location input bar at the top of the viewer.
Zooming in and out
You can zoom into and out of a genomic region by:
- Click the (+) and (-) magnifying glass icons at the top of the viewer.
- Double-clicking on a region will zoom in one level
Selecting Tracks
Use the track selector menu on the right of the viewer to add or remove tracks to the current view.
What do the colors and glyphs mean
See this page: GenomeView examples