Syntenic dotplot: Difference between revisions
No edit summary |
No edit summary |
||
| Line 30: | Line 30: | ||
| Insertion of IS3 and IS2 sequence | | Insertion of IS3 and IS2 sequence | ||
| Pseudogene made in DH10B by IS sequence and transposon insertion<br> | | Pseudogene made in DH10B by IS sequence and transposon insertion<br> | ||
| Possible insertion in | | Possible insertion in REL606 as evidenced by direct repeats.<br> | ||
| [http://tinyurl.com/yedn5tb http://tinyurl.com/yedn5tb]<br> | | [http://tinyurl.com/yedn5tb http://tinyurl.com/yedn5tb]<br> | ||
|- | |- | ||
| Line 157: | Line 157: | ||
|- | |- | ||
| | | | ||
13a. Deletion | 13a. Deletion | ||
13b, Insertion<br> | 13b, Insertion<br> | ||
13c. Insertion | 13c. Insertion | ||
and deletion<br> | and deletion<br> | ||
| Line 206: | Line 206: | ||
|- | |- | ||
| | | | ||
14a. Insertion | 14a. Insertion | ||
14b. Insertion | 14b. Insertion | ||
14c. Deletion | 14c. Deletion | ||
| | | | ||
Revision as of 21:20, 16 October 2009
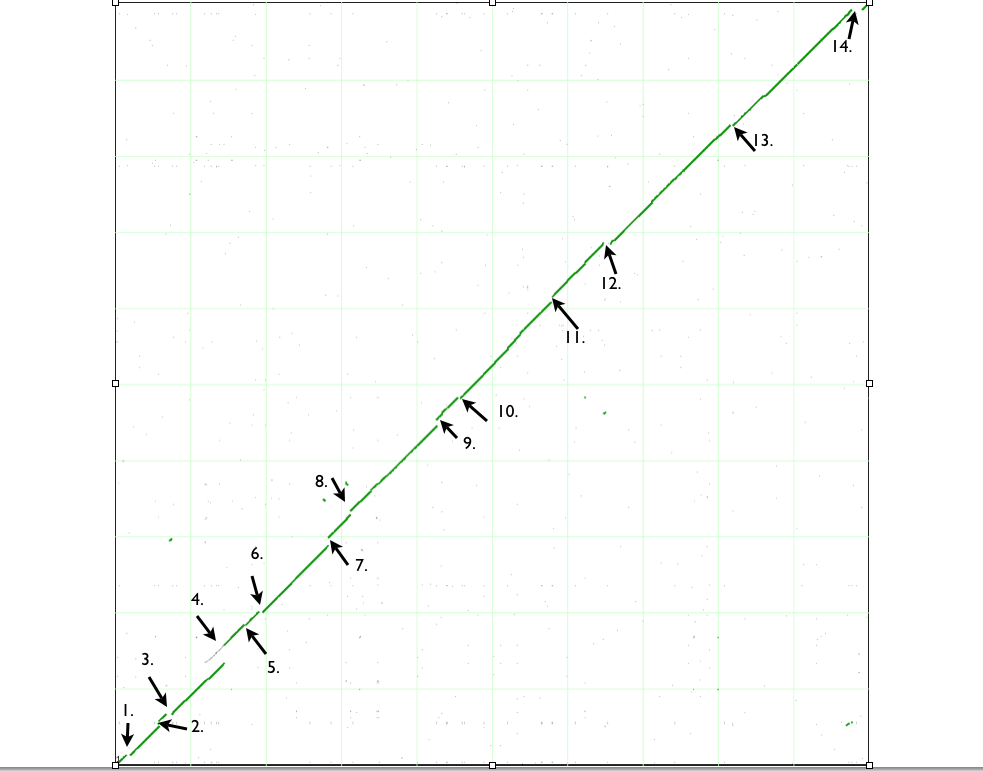
| Variation type |
Difference in strain B REL606 |
Difference in strain K-12 DH10B |
Evidence |
Notes |
Link leading to GEvo |
| 1. Deletion |
none |
Deletion of ~18 genes including DNA pol II, genes in metabolic pathway, thiamine ABC transporter |
pseudogenes in DH10B at deletion site. |
Possible additional insertion in DH10B as evidenced by pseudogenes of yabP, RNA pol associated helicase and FruR, that are not present in REl606 |
tinyurl.com/yexrzpb |
| 2. Insertion |
Insertion of IS1 sequence and a "predicted" transposase |
Insertion sequences and Prophage CP46 DNA insertion | Prophage specific genes found |
Prophage DNA insertion created pseudogenes in K-12 DH10B |
tinyurl.com/yd2quy7 |
| 3. Deletion in B strain or Insertion in K12 strain |
Insertion of IS1 sequence. Insertion of ~15 genes including lac operon and other metabolic enzymes genes | Insertion of IS3 and IS2 sequence | Pseudogene made in DH10B by IS sequence and transposon insertion |
Possible insertion in REL606 as evidenced by direct repeats. |
http://tinyurl.com/yedn5tb |
| 4. Insertion in B strain and DNA duplication event in K12. |
Prophage DNA and transposase insertion |
Recent DNA duplication event | 100% identity between paralogs and ~98% identity between syntenic region in REL606 |
Possible phage DNA insertion in REL606 as "hypothetical protein" genes were found near putative prophage tail component gene in REL606. |
tinyurl.com/yea8bu6 |
| 5. Insertion |
Bacteriophage DNA insertion |
IS2 sequence insertion |
Pseudogenes at IS2 insertion site in DH10B. Phage specific genes were found in REL606 |
Possible phage DNA insertion in REL606 as "Hypothetical proteins" were found near phage specific genes |
tinyurl.com/yevlb2w |
| 6. Insertion |
Prophage DNA insertion |
none |
Phage specific genes were found in REL606 |
none |
tinyurl.com/ybokuag |
| 7. Insertion |
none | Prophage DNA insertion | Phage specific genes found in DH10B |
none |
tinyurl.com/yaxlh7o |
| 8. Insertion and deletion |
Transposon insertions and deletion of phenylacetic acid degradation genes |
IS and Rac prophage DNA insertion | Phage specific genes found in DH10B. IS insertions in REL606 might have created direct repeats and facilitated excision of phenylacetic acid degradation genes. |
Rac prophage DNA disrupted by transposon insertion in DH10B | tinyurl.com/yccbmsq |
|
9a. Insertion 9b. Insertion 9c. Insertion 9d. Insertion |
9a. none 9b. Insertion of ISI transposon 9c. none 9d. IS2 insertion |
9a. Insertion of IS5 sequence 9b. none 9c. Insertion of ABC transporter, flagella encoding genes and few other enzymes 9d. none |
9a. none 9b. none 9c. Inserted DNA segment in DH10B is bordered by direct repeats at both ends. 100% identity was found between the two repeats. 9d. none |
9a. none 9b. none 9c.DR indicates transposon insertion in DH10B. 9d. none |
|
| 10. Insertion and deletion |
Bacteriophage DNA insertion and IS1 transposon insertion. Deletion of metal resistance genes |
Insertion of pili associated genes | Insertion of pili associated genes in DH10B as evidenced by their different GC codon | No IR were found as to indicate insertion of genes conferring metal resistance in DH10B. Possible deletion in REL606 of these genes prior to IS1 transposon insertion. | |
| 11. Insertion |
IS1 insertion followed by insertion of ParB family protein genes and recombinase. |
CP4-57 prophage DNA insertion |
Different GC content of ParB family protein and recombinase gene indicates that these genes were acquired by exogenous DNA such as plasmid possibly by homologous crossover as no IR were found. | IS1 insertion probably occured independent of integration of other genes in REL606. | http://tinyurl.com/ycdog2c |
| 12. Insertion, deletion and Inversion |
IS1 insertion. Insertion of pyrophosphorylase and "hypothetical protein" genes | IS5 and IS10 transposon insertion. Inversion of ornithine decarboxylase, M-type protein and bifunctional prepilin peptidase/methylase. Deletion of saframycin synthetase, capsule related genes, bio-film formation genes, anti-toxin system and type II secretory apparatus genes. |
Insertion of pyrophosphorylase and "hypthetical protein' genes in REL606 as evidenced by their different GC content Inversion in DH10B as evidenced by IR of IS10 transposon. Deletion of several genes in DH10B is evidenced by IS5 transposase transactivator. |
Insertion of IS5 trans-activator transposase indicates possible deletion of several genes in DH10B. Also, no evidence of insertion in REL606 was found such as different GC content or DR |
http://tinyurl.com/ycoagxh |
|
13a. Deletion 13b, Insertion 13c. Insertion and deletion |
13a. none 13b. Insertion of IS1, waaL and waaV genes 13c. Insertion of IS30 transposon and several 'hypothetical protein" genes. |
13a. Deletion of putative adhesin 13b. Insertion of ~5 rfa genes. 13c. Deletion of ShiA-like and TrbC-like protein genes |
13a. Insertion of HEAT repeat containing lyase at the site of deletion in DH10B may have created pseudogene of adhensin which later got deleted 13b. Insertion in both is evideneced by different GC content of the genes. 13c. Insertion in REL606 is evidenced by different GC content of genes.
|
13a.Insertion of HEAT repeat containing lyase is evidenced by its different GC content. Insertion may have created pseudogene of adhesin which later got deleted,in REL606 as evidenced by different GC content of capsule related protein. 13b. IS1 insertion created pseudogene. 13c. Pseudogene in REL606 and different GC content of genes indicate possible insertion. No DRs were found as to indicate insertion of ShiA-like and TrbC-like gene therefore deletion of these genes in DH10B may have occured |
13a. tinyurl.com/yjojy53 13b.http://tinyurl.com/yj2yg5s 13c. tinyurl.com/yjzdyum |
|
14a. Insertion 14b. Insertion 14c. Deletion |
14a. Inserion of several transposons and secondary glycine betaine transporter 14b. Insertion of ~15 genes 14c. none |
14a.Insertion of several transposons. Insertion of Kple2 phage-like element 14b. none 14c. Deletion of ~15 genes.
|
14a. Insertion of transposons has integreted genes of different GC contents, 14b. Insertion in REL606 is evidenced by IR flanking the DNA segment containing several genes. 14c. Deletion in DH10B is evidenced by insertion of IS10R which may have facilitated excision of DNA by forming DRs |
14a. none 14b.Pseudogene created at the site of insertion in DH10B. 14c. none |
14a. tinyurl.com/yzyvunx 14b. tinyurl.com/yly2b6u 14c. tinyurl.com/yfhhsk6 |