Source code: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
|||
| Line 16: | Line 16: | ||
#[[GeLo | CoGePedia page]] | #[[GeLo | CoGePedia page]] | ||
#[http://synteny.cnr.berkeley.edu/CoGe/example/GeLo/example_CoGe_Genome_Browser.tar.gz Tarball] | #[http://synteny.cnr.berkeley.edu/CoGe/example/GeLo/example_CoGe_Genome_Browser.tar.gz Tarball] | ||
#[http://synteny.cnr.berkeley.edu/CoGe/example/GeLo/ Example implementation] using the | #[http://synteny.cnr.berkeley.edu/CoGe/example/GeLo/ Example implementation] using the [http://code.google.com/p/genome-browser/downloads/list OpenGenes Genome Browser]. | ||
Revision as of 19:02, 30 November 2009
CoGe's software is divided into several components:
- Database (mysql)
- Database API (perl)
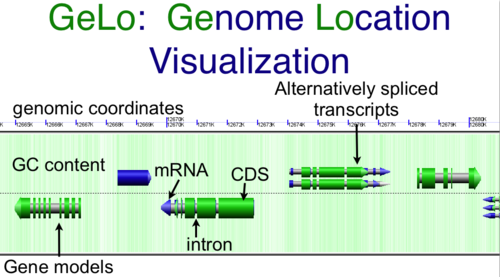
- Genome Location Visualization: GeLo (perl)
- Website and applications (perl, javascript, python, flash, HTML)
- Other stuff: alignment algorithms, etc.
As documentation often lags behind development, much of CoGe's software is not in a suitable condition for release. However, once it has been, we will post its code here. Also, we will be happy to provide any code as requested and do our best to answer any questions you may have about it.
Download CoGe's genome visualization library GeLo