How to extract genomic features: Difference between revisions
Jump to navigation
Jump to search
| Line 7: | Line 7: | ||
===Find your genome of interest in [[OrganismView]]=== | ===Find your genome of interest in [[OrganismView]]=== | ||
[[Image:OrganismView Example Search.png|thumb| | [[Image:OrganismView Example Search.png|thumb|400px|right|Searching for the organism "Arabidopsis thaliana" in OrganismView. Version 9 has been selected containing unmasked genomic sequence. Dataset ch1.xml and chromosome 1 have been selected.]] | ||
This option should be used if you know the organism, chromosome, and nucleotide position of the genomic region in which you are interested. | This option should be used if you know the organism, chromosome, and nucleotide position of the genomic region in which you are interested. | ||
#Go to [[OrganismView ]] and search for your organism by name by typing part of its name in the name search box, or part of the [[organism description]] in the description box. | #Go to [[OrganismView ]] and search for your organism by name by typing part of its name in the name search box, or part of the [[organism description]] in the description box. | ||
Revision as of 05:40, 2 January 2010
Overview
Extracting genomic features from a genomic region in a genome of interest is easy using GenomeView. All you need to do is:
- Find your genome of interest in OrganismView or your genomic feature in FeatView
- Visualize the genomic region of interest in GenomeView
- Select and export a genomic region from OrganismView for automatic feature extraction using FeatList
- Select the features of interest and send them to FastaView to get their sequences in fasta format
Find your genome of interest in OrganismView
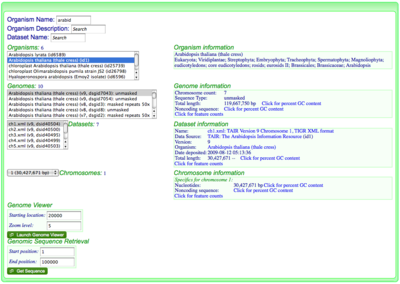
This option should be used if you know the organism, chromosome, and nucleotide position of the genomic region in which you are interested.
- Go to OrganismView and search for your organism by name by typing part of its name in the name search box, or part of the organism description in the description box.
- Select the correct organism from the organism list
- Select the appropriate genome from the list of genomes for that organism
- If needed, select the appropriate dataset and chromosome for the genome