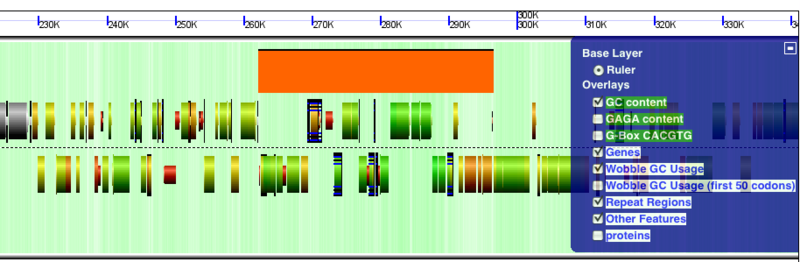
GenomeView examples: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
[[Image:GeLo-example.png|thumb|800px|center|Overview of [[GenomeView]]'s visualization using the [[GeLo]] genomic visualization library. Background of chromosome is colored green for GC rich regions and while for AT rich regions. Gene models are drawn as composite colored arrows such that gray is the extent of the gene, blue is mRNA, and green is protein coding [[CDS]] sequence. Overlapping genomic features are detected and expanded to show alternatively spiced transcripts]] | [[Image:GeLo-example.png|thumb|800px|center|Overview of [[GenomeView]]'s visualization using the [[GeLo]] genomic visualization library. Background of chromosome is colored green for GC rich regions and while for AT rich regions. Gene models are drawn as composite colored arrows such that gray is the extent of the gene, blue is mRNA, and green is protein coding [[CDS]] sequence. Overlapping genomic features are detected and expanded to show alternatively spiced transcripts]] | ||
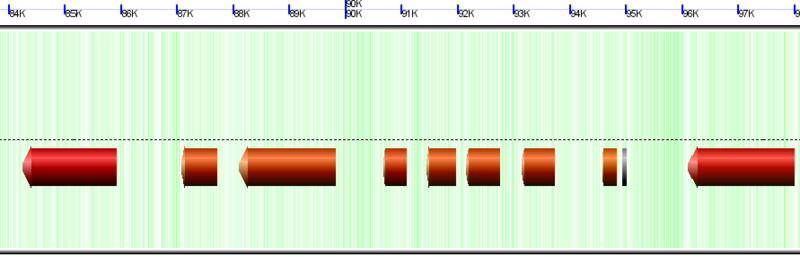
===Background=== | ===Chromosome Background=== | ||
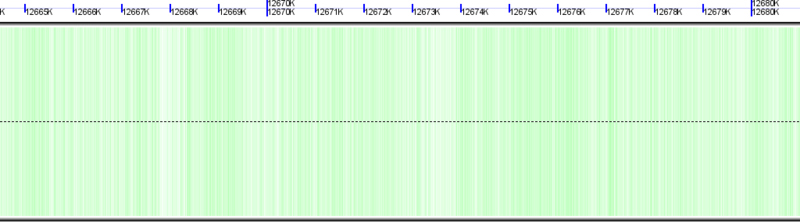
[[Image:GenomeView-background.png|thumb|800px|center|Background of chromosome colored green to show GC rich regions and white to show AT rich regions]] | [[Image:GenomeView-background.png|thumb|800px|center|Background of chromosome colored green to show GC rich regions and white to show AT rich regions]] | ||
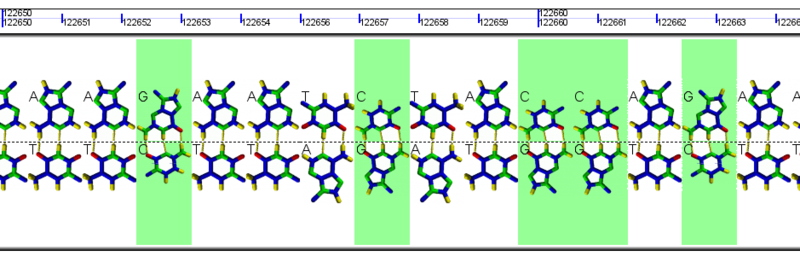
[[Image:GeLo-nucleotides.png|thumb|800px|center|Example GeLo graphics using images of nucleotides in the background. These are also colored green for GC and white for AT]] | [[Image:GeLo-nucleotides.png|thumb|800px|center|Example GeLo graphics using images of nucleotides in the background. These are also colored green for GC and white for AT]] | ||
Revision as of 03:57, 21 February 2010
Overview
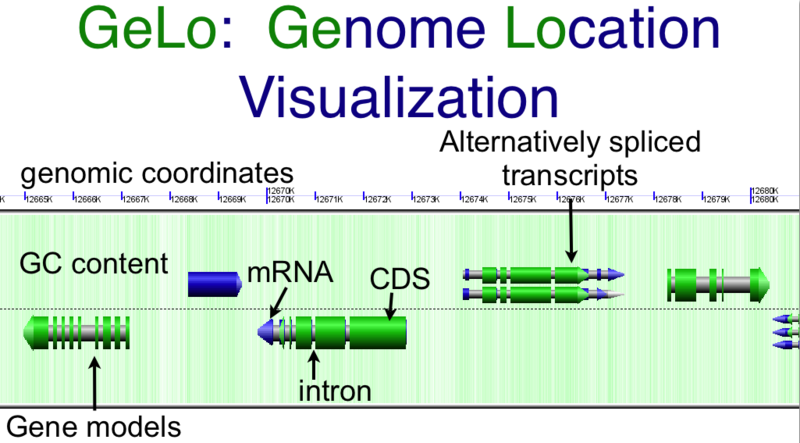
Chromosome Background
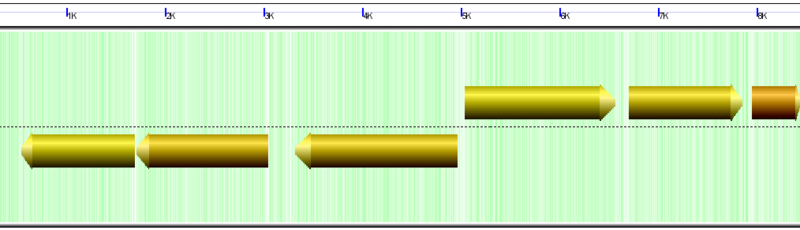
Genes
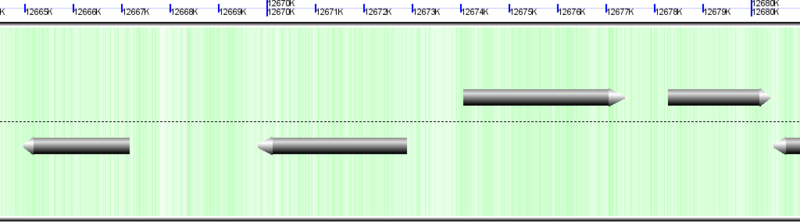
mRNAs
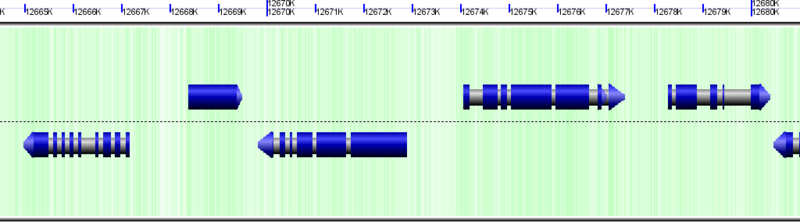
CDSs
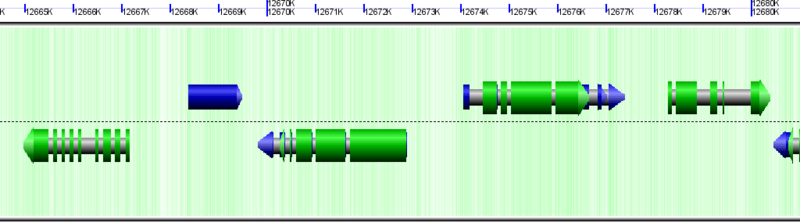
Unsequenced regions
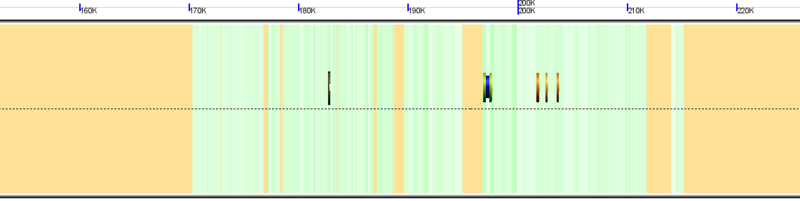
Masked regions
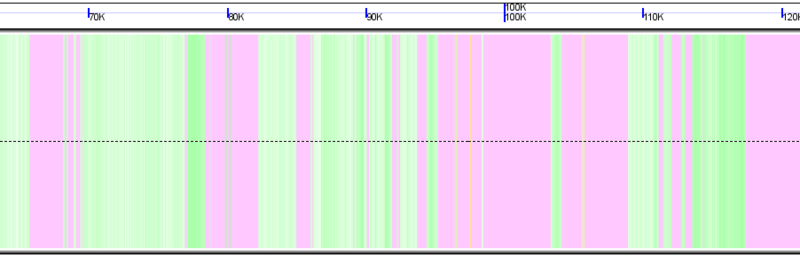
rRNA genes
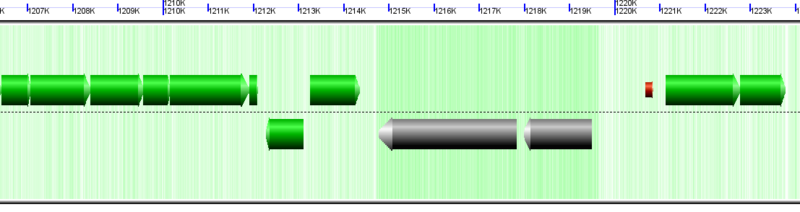
Pseudogenes
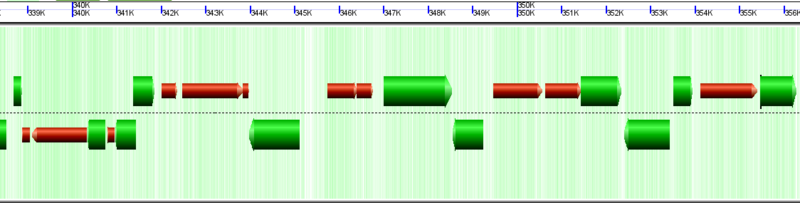
Wobble GC content filter
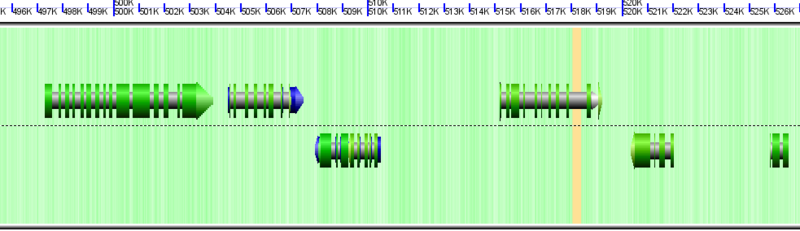
GEvo Links
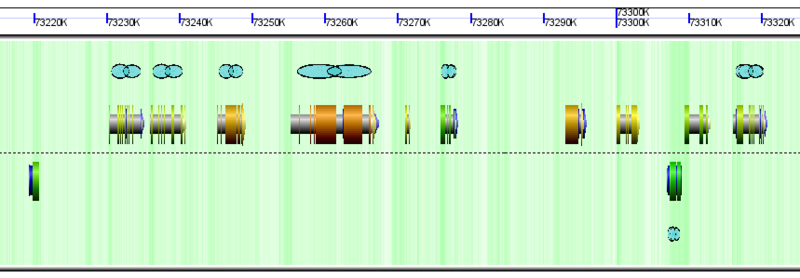
Repeat Regions
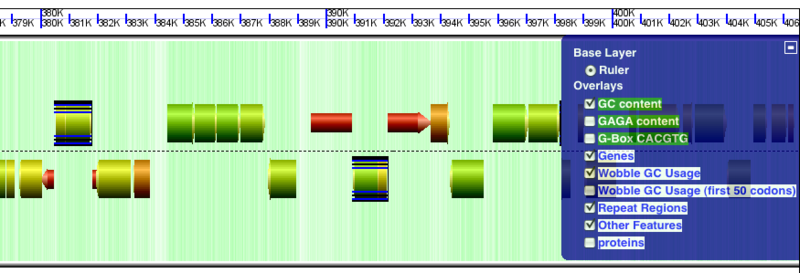
Other Features