Fractionation: Difference between revisions
No edit summary |
No edit summary |
||
| Line 11: | Line 11: | ||
Fractionation is a natural process where genomic regions that contain genes, [[regulatory elements]], or other [[genomic features]] are lost over evolutionary time. This process is especially prevalent in plant genomes with a history of [[polyploidy]] events. After the duplication of a genomic region, or the entire genome as is the case with [[tetraploidy]], most of the duplicated genomic features are lost from one [[homeolog]] or the other. This is assumed to happen because there is generally no selection for the retention of these features in duplicate. (However, it is important to note that there are several classes of genomic features that are selected to be retained following genome duplication events. As example of such a class are [[transcription factor]] genes.) | Fractionation is a natural process where genomic regions that contain genes, [[regulatory elements]], or other [[genomic features]] are lost over evolutionary time. This process is especially prevalent in plant genomes with a history of [[polyploidy]] events. After the duplication of a genomic region, or the entire genome as is the case with [[tetraploidy]], most of the duplicated genomic features are lost from one [[homeolog]] or the other. This is assumed to happen because there is generally no selection for the retention of these features in duplicate. (However, it is important to note that there are several classes of genomic features that are selected to be retained following genome duplication events. As example of such a class are [[transcription factor]] genes.) | ||
When [[conserved noncoding sequences]] ([[CNSs]]) are fractionated, this is evidence that the [[cis-regulation]] of their nearby genes is being [[subfuncationalized]]. Since the regulatory function of CNSs is inferred, we do not know how they may be regulating gene function (e.g. [[enhancers]], [[repressors]]) | When [[conserved noncoding sequences]] ([[CNSs]]) are fractionated, this is evidence that the [[cis-regulation]] of their nearby genes is being [[subfuncationalized]]. Since the regulatory function of CNSs is inferred, we do not know how they may be regulating gene function (e.g. [[enhancers]], [[repressors]]). | ||
Revision as of 19:23, 26 February 2010
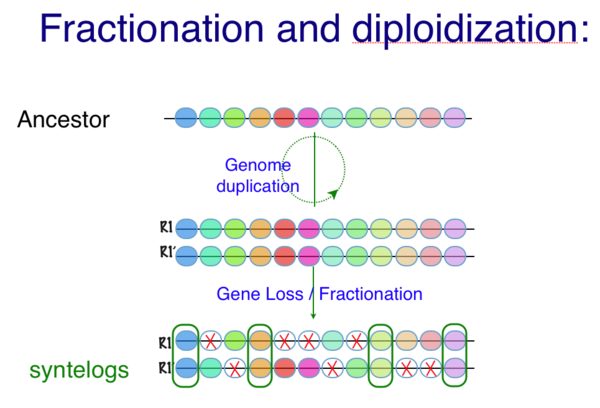

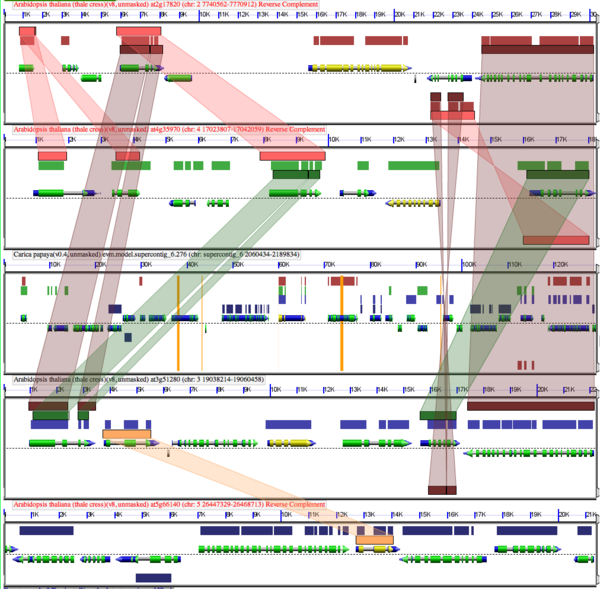
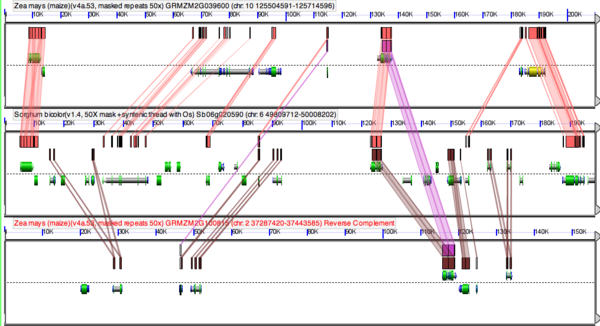
Following whole genome duplication (WGD) events, fractionation is the process of gene loss from one homeologous genomic region or its partner region. Over evolutionary time, this returns a genome to its diploid state in terms of overall gene content. Thus, fractionation is part of the diploidization process. However, the overall structure of the genome will be vastly different as genes are lost from one homeologous region or the other, resulting in genes having different neighboring genes. Also, some families of genes are retained preferentially following WGD events.
Fractionation is a natural process where genomic regions that contain genes, regulatory elements, or other genomic features are lost over evolutionary time. This process is especially prevalent in plant genomes with a history of polyploidy events. After the duplication of a genomic region, or the entire genome as is the case with tetraploidy, most of the duplicated genomic features are lost from one homeolog or the other. This is assumed to happen because there is generally no selection for the retention of these features in duplicate. (However, it is important to note that there are several classes of genomic features that are selected to be retained following genome duplication events. As example of such a class are transcription factor genes.)
When conserved noncoding sequences (CNSs) are fractionated, this is evidence that the cis-regulation of their nearby genes is being subfuncationalized. Since the regulatory function of CNSs is inferred, we do not know how they may be regulating gene function (e.g. enhancers, repressors).