Splitting maize genome: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image:Reconstructingchromosomes.png|From a maize-sorghum dot plot. "A" represents one ortholog of sorghum chromosome 1 in maize. B marks the only possible way to reconstruct the second copy: combining parts of maize chromosomes 5 and 9. This logic allows us to reconstruct pairs of psuedo-sorghum chromosomes out of the maize genome.]] | [[Image:Reconstructingchromosomes.png|left|thumb|From a maize-sorghum dot plot. "A" represents one ortholog of sorghum chromosome 1 in maize. B marks the only possible way to reconstruct the second copy: combining parts of maize chromosomes 5 and 9. This logic allows us to reconstruct pairs of psuedo-sorghum chromosomes out of the maize genome.]] | ||
The ancestor of modern maize/corn went through a [[Whole genome duplication]] sometime between 5 and 12 million years ago (after the maize and sorghum lineages split). Using the chromosomes of the sorghum genome as an outgroup we can accurately identity which parts of each chromosome are represented by which parts of the maize genome. | The ancestor of modern maize/corn went through a [[Whole genome duplication]] sometime between 5 and 12 million years ago (after the maize and sorghum lineages split). Using the chromosomes of the sorghum genome as an outgroup we can accurately identity which parts of each chromosome are represented by which parts of the maize genome. | ||
The logic used to reconstruct maize chromosome pairs doesn't allow us to assign those chromosomes into two unique psuedo-genomes. Reconstructed chromosomes were assigned to maize1 or maize2 based on the direction of biased gene loss. Maize1 consists of the reconstructed chromosomes from which fewer genes have been lost since the maize whole genome duplication and maize2 of the reconstructed chromosomes from which more genes have been lost (as determined by comparison to the sorghum outgroup). That's ALL it means. (figure 2) | The logic used to reconstruct maize chromosome pairs doesn't allow us to assign those chromosomes into two unique psuedo-genomes. Reconstructed chromosomes were assigned to maize1 or maize2 based on the direction of biased gene loss. Maize1 consists of the reconstructed chromosomes from which fewer genes have been lost since the maize whole genome duplication and maize2 of the reconstructed chromosomes from which more genes have been lost (as determined by comparison to the sorghum outgroup). That's ALL it means. (figure 2) | ||
Revision as of 03:20, 6 March 2010
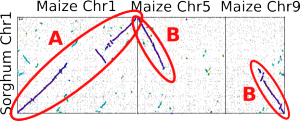
The ancestor of modern maize/corn went through a Whole genome duplication sometime between 5 and 12 million years ago (after the maize and sorghum lineages split). Using the chromosomes of the sorghum genome as an outgroup we can accurately identity which parts of each chromosome are represented by which parts of the maize genome.
The logic used to reconstruct maize chromosome pairs doesn't allow us to assign those chromosomes into two unique psuedo-genomes. Reconstructed chromosomes were assigned to maize1 or maize2 based on the direction of biased gene loss. Maize1 consists of the reconstructed chromosomes from which fewer genes have been lost since the maize whole genome duplication and maize2 of the reconstructed chromosomes from which more genes have been lost (as determined by comparison to the sorghum outgroup). That's ALL it means. (figure 2)