Syntenic path assembly: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
<gallery widths=500px heights=500px caption= | <gallery widths=500px heights=500px caption=''"Syntenic Path Assembly in SynMap of E. coli''"''> | ||
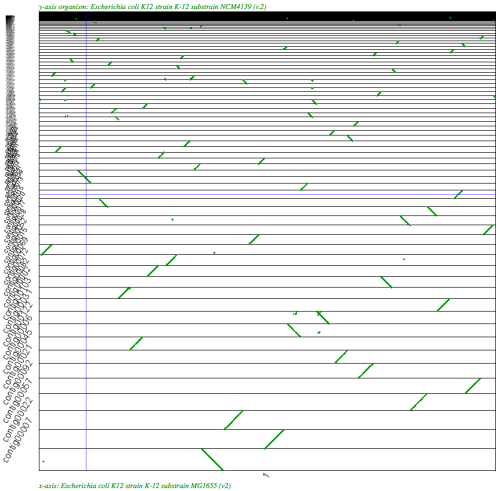
File:Screen shot 2011-02-18 at 1.24.54 PM.png|[[SynMap]] analysis of a WGS assembly of a strain of E. coli K12 (y-axis) to a reference assembly (x-axis). Results may be regenerated at http://genomevolution.org/r/2k70 | File:Screen shot 2011-02-18 at 1.24.54 PM.png|[[SynMap]] analysis of a WGS assembly of a strain of E. coli K12 (y-axis) to a reference assembly (x-axis). Results may be regenerated at http://genomevolution.org/r/2k70 | ||
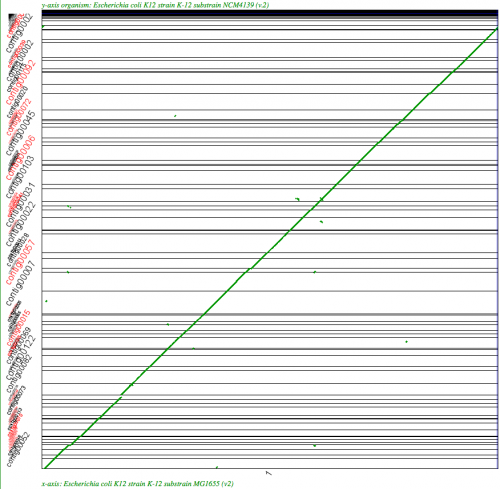
File:Screen shot 2011-02-18 at 1.27.34 PM.png|[[SynMap]] analysis of a WGS assembly of a strain of E. coli K12 (y-axis) to a reference assembly (x-axis) using SynMap's Syntenic Path Assembly to order contigs. Results may be regenerated at: http://genomevolution.org/r/2k71 | File:Screen shot 2011-02-18 at 1.27.34 PM.png|[[SynMap]] analysis of a WGS assembly of a strain of E. coli K12 (y-axis) to a reference assembly (x-axis) using SynMap's Syntenic Path Assembly to order contigs. Results may be regenerated at: http://genomevolution.org/r/2k71 | ||
Revision as of 20:11, 18 February 2011
-
SynMap analysis of a WGS assembly of a strain of E. coli K12 (y-axis) to a reference assembly (x-axis). Results may be regenerated at http://genomevolution.org/r/2k70
-
SynMap analysis of a WGS assembly of a strain of E. coli K12 (y-axis) to a reference assembly (x-axis) using SynMap's Syntenic Path Assembly to order contigs. Results may be regenerated at: http://genomevolution.org/r/2k71
The Syntenic Path Assembly is an option in SynMap to do a reference genome assembly of contigs using synteny to determine the order and orientation of the contigs.