Genomic Rearrangement Analysis: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
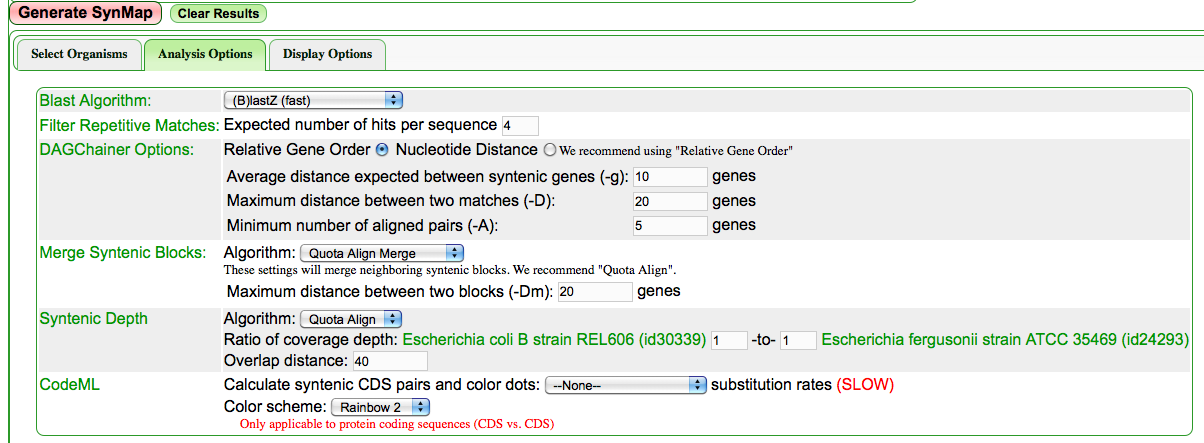
[[Image:Screen shot 2011-03-11 at 7.58.21 AM.png|Configuring a SynMap analysis to use [[Quota align]] with a syntenic depth of 1:1.]] | [[Image:Screen shot 2011-03-11 at 7.58.21 AM.png|frame|Configuring a SynMap analysis to use [[Quota align]] with a syntenic depth of 1:1.]] | ||
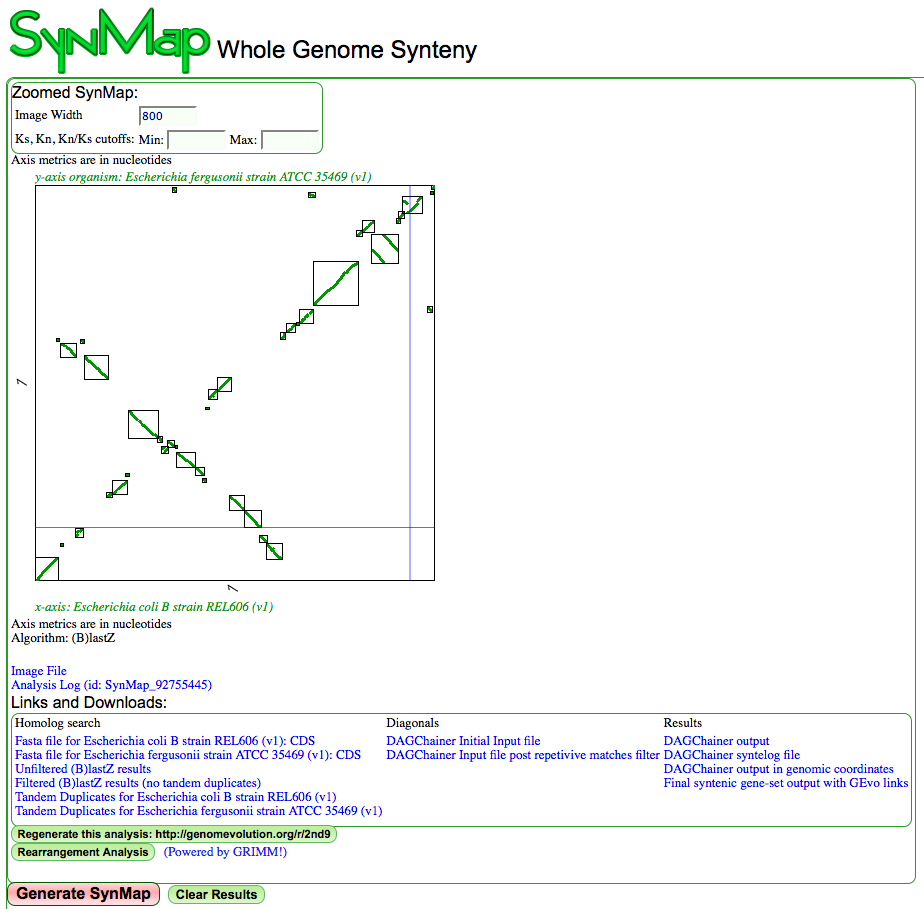
[[Image:Screen shot 2011-03-11 at 7.58.04 AM.png| | [[Image:Screen shot 2011-03-11 at 7.58.04 AM.png|frame|Syntentic dotplot between two species of Escherichia. X-axis: coli B strain REL606; Y-axis: fergusonii strain ATCC 35469. [[Syntenic dotplot]] has boxes drawn around syntenic regions and there is a link to GRIMM for rearrangement analysis. The link to GRIMM will only show if the [[quota align]] algorithm has been set and set to a 1:1 ratio. Results may be regenerated at http://genomevolution.org/r/2nd7]] | ||
Revision as of 13:53, 11 March 2011