Inversion: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image:SynMap-inversion.png|thumbnail|right|500 px|[http://synteny.cnr.berkeley.edu/CoGe//diags/Arabidopsis_lyrata/Arabidopsis_thaliana_thale_cress/html/3068_8.CDS-CDS.blastn.dag_geneorder_D20_g10_A5.scaffold_4-2.w600.html|[http://synteny.cnr.berkeley.edu/CoGe//diags/Arabidopsis_lyrata/Arabidopsis_thaliana_thale_cress/html/3068_8.CDS-CDS.blastn.dag_geneorder_D20_g10_A5.scaffold_4-2.w600.html]]] | |||
[[Image:GEvo-inversion.png|thumbnail|right|500 px|[http://toxic.berkeley.edu/CoGe/GEvo.pl?prog=blastz;spike_len=0;accn1=fgenesh2_kg.4__765__AT2G28060.1;fid1=30750039;dsid1=39129;dsgid1=3068;gstid1=1;chr1=scaffold_4;dr1up=830865;dr1down=1073591;gbstart1=1;gblength1=2332;mask1=non-cds;do1=1;accn2=AT2G28060;fid2=20219657;dsid2=35172;dsgid2=8;gstid2=1;chr2=2;dr2up=334450;dr2down=533528;gbstart2=1;gblength2=1591;mask2=non-cds;do2=2;num_seqs=2;hsp_overlap_limit=0;hsp_size_limit=0]]] | [[Image:GEvo-inversion.png|thumbnail|right|500 px|[http://toxic.berkeley.edu/CoGe/GEvo.pl?prog=blastz;spike_len=0;accn1=fgenesh2_kg.4__765__AT2G28060.1;fid1=30750039;dsid1=39129;dsgid1=3068;gstid1=1;chr1=scaffold_4;dr1up=830865;dr1down=1073591;gbstart1=1;gblength1=2332;mask1=non-cds;do1=1;accn2=AT2G28060;fid2=20219657;dsid2=35172;dsgid2=8;gstid2=1;chr2=2;dr2up=334450;dr2down=533528;gbstart2=1;gblength2=1591;mask2=non-cds;do2=2;num_seqs=2;hsp_overlap_limit=0;hsp_size_limit=0]]] | ||
An inversion happens when a genomic region flips its orientation in a genome such that the 5' and 3' ends change places. There are several ways to visualize a genomic inversion. | An inversion happens when a genomic region flips its orientation in a genome such that the 5' and 3' ends change places. There are several ways to visualize a genomic inversion. The two images at the right use CoGe to visualize an inversion. The top image is a syntenic dotplot between | ||
Revision as of 18:33, 21 July 2009
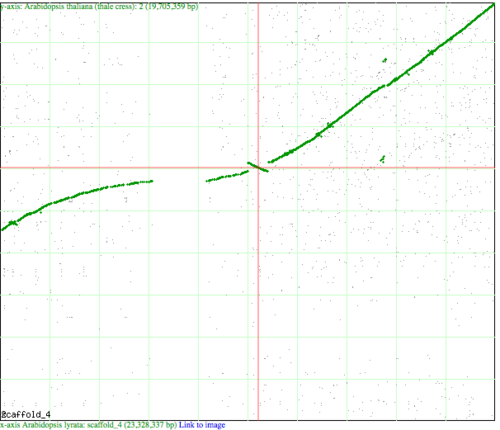

An inversion happens when a genomic region flips its orientation in a genome such that the 5' and 3' ends change places. There are several ways to visualize a genomic inversion. The two images at the right use CoGe to visualize an inversion. The top image is a syntenic dotplot between