Plant paleopolyploidy: Difference between revisions
| Line 31: | Line 31: | ||
===3 Arabidopsis beta=== | ===3 Arabidopsis beta=== | ||
As of yet not a single lineage has been identified in which the beta tetraploidy (naming conventions from Bowers et al 2003<ref name="bowers" />) is the most recent whole genome duplication. | As of yet not a single lineage has been identified in which the beta tetraploidy (naming conventions from Bowers et al 2003<ref name="bowers" />) is the most recent whole genome duplication. Despite what the image above might indicate, this duplication is significantly older than arabidopsis alpha, however precise dating is difficult given the acceleration of synonymous substitution rate in the arabidopsis lineage. | ||
===4 Brassica hexaploidy=== | ===4 Brassica hexaploidy=== | ||
| Line 41: | Line 41: | ||
When the genome of poplar was released back in 2006, researchers announced that they had identified a new ancient whole genome duplication<ref name="poplar genome">Tuskan GA et al (2006) "The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray)" ''Science'' DOI: [http://dx.doi.org/10.1126/science.1128691 10.1126/science.1128691]</ref>. Poplar retains around 8000 pairs of duplicated genes. | When the genome of poplar was released back in 2006, researchers announced that they had identified a new ancient whole genome duplication<ref name="poplar genome">Tuskan GA et al (2006) "The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray)" ''Science'' DOI: [http://dx.doi.org/10.1126/science.1128691 10.1126/science.1128691]</ref>. Poplar retains around 8000 pairs of duplicated genes. | ||
=== | ===7 Apple tetraploidy=== | ||
= | The genome paper of apple also discussed an ancient whole genome duplication identified in that linage which they estimated to be >50 million years old <ref name="apple genome">Valasco R et al (2010) "The genome of the domesticated apple (Malus × domestica Borkh.)" ''Nature Genetics'' DOI: [http://dx.doi.org/10.1038/ng.654 10.1038/ng.654]</ref> | ||
===8 Soybean tetraploidy=== | ===8 Soybean tetraploidy=== | ||
The relatively recent whole genome duplication in soybean was long suspected based a number of different forms of analysis, from analysis of Ks peaks<ref>Schlueter JA et al (2004) "Mining EST databases to resolve evolutionary events in major crop species." ''Genome'' DOI: [http://dx.doi.org/10.1139/g04-047 10.1139/g04-047]</ref>, phylogenies of individual gene families<ref>Pfeil BE et al (2005) "Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families." ''Systematic Biology'' DOI: [http://dx.doi.org/10.1080/10635150590945359 10.1080/10635150590945359]</ref> and analyzing the [[fractionation]] of individual sequenced regions<ref>Schlueter JA et al (2008) "ractionation of synteny in a genomic region containing tandemly duplicated genes across Glycine max, Medicago truncatula, and Arabidopsis thaliana." ''Journal of Heredity'' DOI: [http://dx.doi.org/10.1093/jhered/esn010 10.1093/jhered/esn010]</ref>. As expected, when the genome of soybean was published in 2010, researchers did identify a recent whole genome duplication (peak ks=0.13, estimated age 13 million years ago). | |||
===9 Legume tetraploidy=== | ===9 Legume tetraploidy=== | ||
The whole genome duplication shared by all legumes was initially inferred by comparison of the medicago and lotus genome assemblies to data from peanut (''Arachis hypogaea'') a basal lineage of that clade<ref>Bertioli DJ et al (2009) "An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes" ''BMC Genomics'' DOI:[http://dx.doi.org/10.1186/1471-2164-10-45 10.1186/1471-2164-10-45].</ref> | |||
===10 Columbine tetraploidy=== | ===10 Columbine tetraploidy=== | ||
Revision as of 19:43, 9 November 2011
The plant lineage, and specifically angiosperms, have a genomic history of repeated whole genome duplication events. This pattern of changing their ploidy level (or number of copies of their genome) is something that happens frequently today, and many domesticated crop plants have been selected with various ploidy levels. While the mechanisms are unknown that permit polyploidy is some lineages such as plants, and not as frequently in others such as mammals [1].
Identifying and characterizing plant paleopolyploidies is ongoing research. They are identified through whole genome comparisons using a combination of the data derived from genomic structure (e.g. syntenic dotplots) and evolutionary distances (e.g. synonymous mutation rates). As such, detecting these events and determining which lineages share what subset are continually changing. The images presented here represent our views right now, but are subject to change. Events that were previously undetected or missed can suddenly be seen with an improved build of a genome or the the sequencing of a fortuitously placed outgroup.
As new genomes become available, and previous genomes are updated, we will continue to improve these figures.
Plant Phylogenetic Tree With WGD Marked
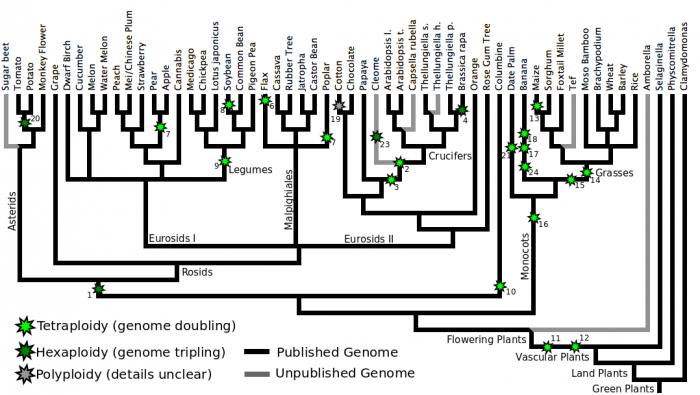
Discussion of individual tetraploidies
1 Eudicot Hexaploidy
Synonyms: Arabidopsis Gamma
This hexaploidy (genome tripling) is shared by the core eudicots (the rosids and asterids), and may be present in additional, basal eudicots, although it will not be possible to reach this conclusion until the genomes of species from basal lineages are sequences -- AND WELL ASSEMBLED!
This whole genome duplication was first identified as the most ancient of three whole genome duplication present in the genome of Arabidopsis thaliana, and assigned the name "gamma" by in 2003 by Bowers et al. [2]:
In this paper, hampered by the fact that only two plant genomes had yet been sequenced, and the ridiculously accelerated rate of base pair substitution in arabidopsis the authors concluded the gamma event was likely shared by both monocots and eudicots and could potentially be as old as the split between gymnosperms and flowering plants 300 million years ago.
With the publication of the grape genome in 2007[3] which has not experienced any duplications since the eudicot hexaploidy and doesn't show the same acceleration of nucleotide substitutions, it became possible to conclude that the eudicot hexaploidy was NOT shared with monocots and was shared by all rosids.
More recent work in the asterids[4] indicate that this highly successful eudicot clade share the same ancient whole genome duplication.
2 Arabidopsis alpha
The alpha tetraploidy of arabidopsis was first given that name in Bowers et al 2003[2]. It is shared by most or all of the crucifers (family Brassicaceae).
3 Arabidopsis beta
As of yet not a single lineage has been identified in which the beta tetraploidy (naming conventions from Bowers et al 2003[2]) is the most recent whole genome duplication. Despite what the image above might indicate, this duplication is significantly older than arabidopsis alpha, however precise dating is difficult given the acceleration of synonymous substitution rate in the arabidopsis lineage.
4 Brassica hexaploidy
The hexaploidy shared by all species in the genus Brassica is well known. So well known, I don't know what the proper citation for the discovery of this whole genome duplication is. If you know, please get in touch so the scientists responsible for this discovery -- however long ago -- can get proper credit.
5 Poplar tetraploidy
When the genome of poplar was released back in 2006, researchers announced that they had identified a new ancient whole genome duplication[5]. Poplar retains around 8000 pairs of duplicated genes.
7 Apple tetraploidy
The genome paper of apple also discussed an ancient whole genome duplication identified in that linage which they estimated to be >50 million years old [6]
8 Soybean tetraploidy
The relatively recent whole genome duplication in soybean was long suspected based a number of different forms of analysis, from analysis of Ks peaks[7], phylogenies of individual gene families[8] and analyzing the fractionation of individual sequenced regions[9]. As expected, when the genome of soybean was published in 2010, researchers did identify a recent whole genome duplication (peak ks=0.13, estimated age 13 million years ago).
9 Legume tetraploidy
The whole genome duplication shared by all legumes was initially inferred by comparison of the medicago and lotus genome assemblies to data from peanut (Arachis hypogaea) a basal lineage of that clade[10]
10 Columbine tetraploidy
11 Flowering plant tetraploidy
12 Seed Plant Tetraploidy
13 Maize Tetraploidy
14 Grass Tetraploidy
15 Monocot Tetraploidy A
16 Monocot Tetraploidy B
- ↑ There is a case of a tetraploid rodent: Gallardo. 2006. Molecular cytogenetics and allotetraploidy in the red vizcacha rat, Tympanoctomys barrerae (Rodentia, Octodontidae) Genomics. 88:2, 214-221. doi:10.1016/j.ygeno.2006.02.010
- ↑ 2.0 2.1 2.2 Bowers JE et al (2003) "Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events." Nature DOI: 10.1038/nature01521
- ↑ Jaillon O et al (2007) "The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla." Nature DOI: 10.1038/nature06148
- ↑ Cenci A et al (2010) "Comparative sequence analyses indicate that Coffea (Asterids) and Vitis (Rosids) derive from the same paleo-hexaploid ancestral genome." Molecular Genetics and Genomics DOI: 10.1007/s00438-010-0534-7
- ↑ Tuskan GA et al (2006) "The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray)" Science DOI: 10.1126/science.1128691
- ↑ Valasco R et al (2010) "The genome of the domesticated apple (Malus × domestica Borkh.)" Nature Genetics DOI: 10.1038/ng.654
- ↑ Schlueter JA et al (2004) "Mining EST databases to resolve evolutionary events in major crop species." Genome DOI: 10.1139/g04-047
- ↑ Pfeil BE et al (2005) "Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families." Systematic Biology DOI: 10.1080/10635150590945359
- ↑ Schlueter JA et al (2008) "ractionation of synteny in a genomic region containing tandemly duplicated genes across Glycine max, Medicago truncatula, and Arabidopsis thaliana." Journal of Heredity DOI: 10.1093/jhered/esn010
- ↑ Bertioli DJ et al (2009) "An analysis of synteny of Arachis with Lotus and Medicago sheds new light on the structure, stability and evolution of legume genomes" BMC Genomics DOI:10.1186/1471-2164-10-45.