Fractionation: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Image:At-Cp-Fractionation.png|thumb|right|500px|Example GEvo analysis of fraction of gene content between 4 syntenic regions of Arabidopsis thaliana and one syntenic region of Carica papaya. Papaya is in the middle with 2 Arabidopsis regions above and below. Since their divergence, Arabidopsis thaliana has had two whole genome duplication events while papaya has had none. After each whole genome duplication event in Arabidopsis, duplicated genes are lost from one homeologous region or the other. Thus, this single region of papaya contains nearly the entire gene content of the four syntenic Arabidopsis regions. Note that some families of genes are retained preferentially following a whole genome duplication event, and that some genes transpose. This analysis can be regenerated at: http://tinyurl.com/o6xnea .]] | [[Image:At-Cp-Fractionation.png|thumb|right|500px|Example GEvo analysis of fraction of gene content between 4 syntenic regions of Arabidopsis thaliana and one syntenic region of Carica papaya. Papaya is in the middle with 2 Arabidopsis regions above and below. Since their divergence, Arabidopsis thaliana has had two whole genome duplication events while papaya has had none. After each whole genome duplication event in Arabidopsis, duplicated genes are lost from one homeologous region or the other. Thus, this single region of papaya contains nearly the entire gene content of the four syntenic Arabidopsis regions. Note that some families of genes are retained preferentially following a whole genome duplication event, and that some genes transpose. This analysis can be regenerated at: http://tinyurl.com/o6xnea .]] | ||
[[Image:At-Cp-Retention.png|thumb|right|500px|Same example GEvo analysis as above, but with retained genes in Arabidopsis highlighted.]] | |||
Following whole genome duplication (WGD) events, fractionation is the process of gene loss from one homeologous genomic region or its partner region. Over evolutionary time, this returns a genome to its diploid state in terms of overall gene content. Thus, fractionation is part of the diploidization process. However, the overall structure of the genome will be vastly different as genes are lost from one homeologous region or the other resulting in genes having different neighboring genes. Also, some families of genes are retained preferentially following WGD events. | Following whole genome duplication (WGD) events, fractionation is the process of gene loss from one homeologous genomic region or its partner region. Over evolutionary time, this returns a genome to its diploid state in terms of overall gene content. Thus, fractionation is part of the diploidization process. However, the overall structure of the genome will be vastly different as genes are lost from one homeologous region or the other resulting in genes having different neighboring genes. Also, some families of genes are retained preferentially following WGD events. | ||
Revision as of 23:31, 4 August 2009
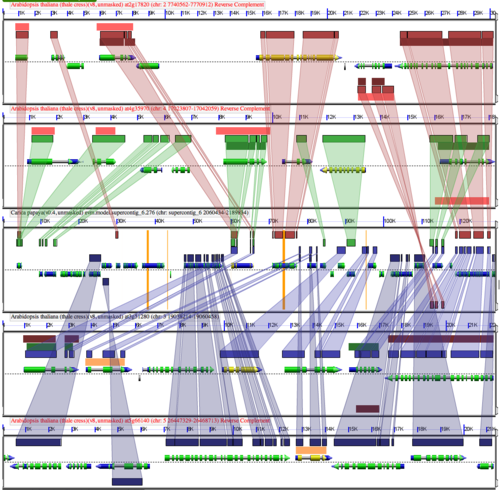
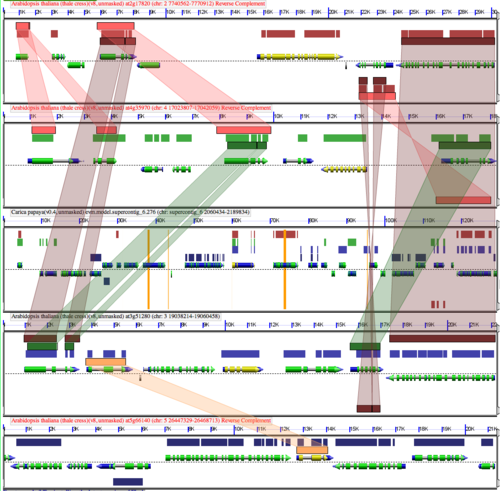
Following whole genome duplication (WGD) events, fractionation is the process of gene loss from one homeologous genomic region or its partner region. Over evolutionary time, this returns a genome to its diploid state in terms of overall gene content. Thus, fractionation is part of the diploidization process. However, the overall structure of the genome will be vastly different as genes are lost from one homeologous region or the other resulting in genes having different neighboring genes. Also, some families of genes are retained preferentially following WGD events.