Syntenic path assembly: Difference between revisions
Jump to navigation
Jump to search
| Line 1: | Line 1: | ||
==Overview== | == Overview == | ||
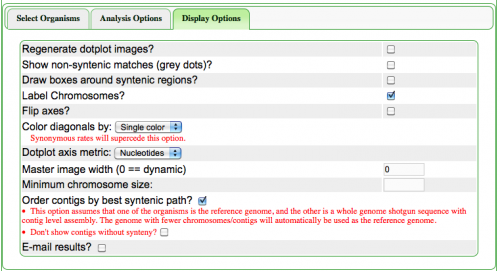
The Syntenic Path Assembly is an option in [[SynMap]] to do a reference genome assembly of contigs using synteny to determine the order and orientation of the contigs. To use this option, select "Order contigs by best syntenic path" under the Display Options tab. When an assembly is generated, you may download the [[Pseudoassembled]] sequence (contigs are joined using 100 "N"s. "N" is the [[Ambiguous nucleotide]] and while it may represent any nucleotide (A, T C, G), this permits the identification of where contigs were "glued" together by this algorithm.). | |||
This algorithm also works quite well in aligning a WGS assembly between distally related organisms. See below for examples. | |||
== Alogrothm == | |||
[[Image:Screen shot 2011-02-18 at 1.38.59 PM.png|thumb | As implemented in SynMap: | ||
[[Image:Screen shot 2011-02-18 at 1.58.23 PM.png|frame|center | |||
#Identify and score each syntenic region | |||
#Assign genome with fewer chromosomes/contigs to be reference | |||
#For each chromosome in reference genome: | |||
# sort all syntenic blocks that match reference genome according to position in reference genome # for ties in position, pick the syntenic block with the greater synteny score # flip contig if syntenic block is reversed | |||
Thus, the tiling looks like: | |||
<pre> | |||
|-----A-----| |-----B-----| |-----C-----| | |||
|-1-||-2-||-3-| |-4-||-5-||-6-| etc., | |||
</pre> | |||
=== Definitions === | |||
*WGS: [[Whole-genome shotgun]] | |||
<br> [[Image:Screen shot 2011-02-18 at 1.38.59 PM.png|thumb|center|500px]] [[Image:Screen shot 2011-02-18 at 1.58.23 PM.png|frame|center]] | |||
==E. coli== | ==E. coli== | ||
Revision as of 21:33, 6 February 2012
Overview
The Syntenic Path Assembly is an option in SynMap to do a reference genome assembly of contigs using synteny to determine the order and orientation of the contigs. To use this option, select "Order contigs by best syntenic path" under the Display Options tab. When an assembly is generated, you may download the Pseudoassembled sequence (contigs are joined using 100 "N"s. "N" is the Ambiguous nucleotide and while it may represent any nucleotide (A, T C, G), this permits the identification of where contigs were "glued" together by this algorithm.).
This algorithm also works quite well in aligning a WGS assembly between distally related organisms. See below for examples.
Alogrothm
As implemented in SynMap:
- Identify and score each syntenic region
- Assign genome with fewer chromosomes/contigs to be reference
- For each chromosome in reference genome:
- sort all syntenic blocks that match reference genome according to position in reference genome # for ties in position, pick the syntenic block with the greater synteny score # flip contig if syntenic block is reversed
Thus, the tiling looks like:
|-----A-----| |-----B-----| |-----C-----| |-1-||-2-||-3-| |-4-||-5-||-6-| etc.,
Definitions
- WGS: Whole-genome shotgun
E. coli
- Syntenic Path Assembly in SynMap of E. coli
-
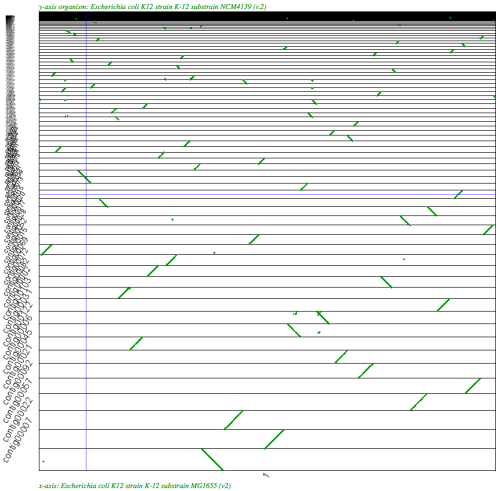
SynMap analysis of a WGS assembly of a strain of E. coli K12 (y-axis) to a reference assembly (x-axis). Results may be regenerated at http://genomevolution.org/r/2k70
-
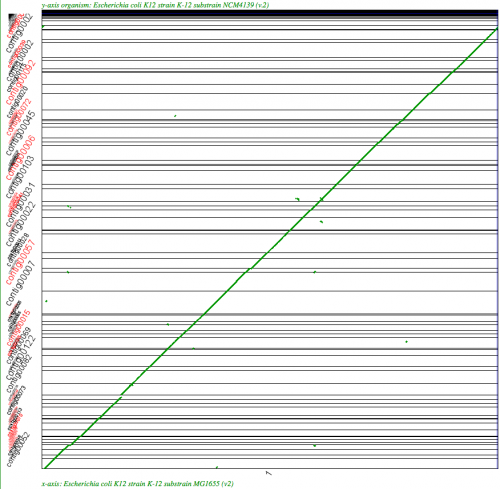
SynMap analysis of a WGS assembly of a strain of E. coli K12 (y-axis) to a reference assembly (x-axis) using SynMap's Syntenic Path Assembly to order contigs. Results may be regenerated at: http://genomevolution.org/r/2k71
Tetraodon (puffer fish): Takifugu rubripes and Tetraodon nigroviridis
-
Tetraodon nigroviridis
-
Takifugo rubripes
-
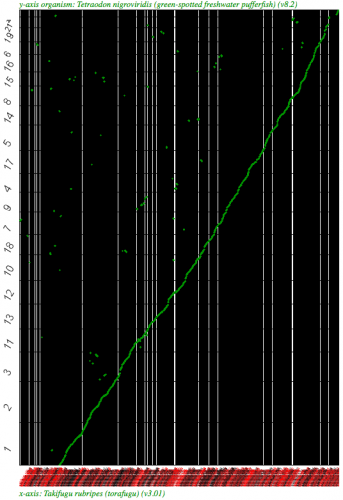
Syntenic dotplot using SynMap between two puffer fish, Takifugu rubripes (x-axis) and Tetraodon nigroviridis (y-axis). (Fugu is a WGS assembly.) Note: the background is colored black due to the number of contigs in the WGS assembly. Each contig/chromosome is visually separated from one another by a vertical or horizontal black line. Results may be regenerated at http://genomevolution.org/r/2k7b
-
Syntneic dotplot using SynMap between two puffer fish, Takifugu rubripes (x-axis) and Tetraodon nigroviridis (y-axis) using the Syntenic Path Assembly option. Fugu is a WGS assembly and fully syntenic converage to Tetraodon is detectable. Results may be regenerated at http://genomevolution.org/r/2k79 . Note: the background is colored black due to the number of contigs in the WGS assembly. Each contig/chromosome is visually separated from one another by a vertical or horizontal black line.
Carnivora: Giant Panda (WGS Assembly) to Dog (reference genome)
-
Dog
-
Syntenic dotplot by SynMap of WGS giant panda genome (x-axis) versus complete dog genome(y-axis). Results may be regenerated at http://genomevolution.org/r/2kbv . Note: the background is colored black due to the number of contigs in the WGS assembly. Each contig/chromosome is visually separated from one another by a vertical or horizontal black line.
-
SynMap Syntneic path assembly of WGS giant panda genome (x-axis) to complete dog genome (y-axis). Results may be regenerated at: http://genomevolution.org/r/2kbt . Note: the background is colored black due to the number of contigs in the WGS assembly. Each contig/chromosome is visually separated from one another by a vertical or horizontal black line.
Arabidopsis ecotypes: Columbia versus Landsberg erecta
-
Syntenic dotplot by [SynMap]. X-axis Columbia; Y-axis Landsberg. Results may be regenerated: http://genomevolution.org/r/3oke
-
Syntenic dotplot by [SynMap]. X-axis Columbia; Y-axis Landsberg. Results may be regenerated: http://genomevolution.org/r/3okf
Phoenix dactylifera L. (date palm) v. Oryza sativa japonica (Rice)
-
Syntenic dotplot by [SynMap]. X-axis Oryza sativa japonica (Rice); Y-axis Phoenix dactylifera L. (date palm). Results may be regenerated: http://genomevolution.org/r/3qjk
-
Syntenic dotplot by [SynMap]. X-axis Oryza sativa japonica (Rice); Y-axis Phoenix dactylifera L. (date palm). Results may be regenerated: http://genomevolution.org/r/3qjn
Cannabis sativa (marijuana) v. Prunus persica (peach)
-
Syntenic dotplot by [SynMap]. X-axis Cannabis sativa (marijuana); Prunus persica (peach). Results may be regenerated: http://genomevolution.org/r/3wz5
-
Syntenic dotplot by [SynMap]. X-axis Cannabis sativa (marijuana); Prunus persica (peach). Results may be regenerated: http://genomevolution.org/r/3wz3







![Syntenic dotplot by [SynMap]. X-axis Columbia; Y-axis Landsberg. Results may be regenerated: http://genomevolution.org/r/3oke](/wiki/images/thumb/1/1d/Master_11022_11937.CDS-genomic.lastz.dag.go_c4_D20_g10_A5.aligncoords.gcoords_ct0.w2000.cs1.csoS.nsd.png/500px-Master_11022_11937.CDS-genomic.lastz.dag.go_c4_D20_g10_A5.aligncoords.gcoords_ct0.w2000.cs1.csoS.nsd.png)
![Syntenic dotplot by [SynMap]. X-axis Columbia; Y-axis Landsberg. Results may be regenerated: http://genomevolution.org/r/3okf](/wiki/images/thumb/1/14/Master_11022_11937.CDS-genomic.lastz.dag.go_c4_D20_g10_A5.aligncoords.gcoords_ct0.w2000.ass.undefined.undefined.cs1.csoS.nsd.png/500px-Master_11022_11937.CDS-genomic.lastz.dag.go_c4_D20_g10_A5.aligncoords.gcoords_ct0.w2000.ass.undefined.undefined.cs1.csoS.nsd.png)
![Syntenic dotplot by [SynMap]. X-axis Oryza sativa japonica (Rice); Y-axis Phoenix dactylifera L. (date palm). Results may be regenerated: http://genomevolution.org/r/3qjk](/wiki/images/thumb/5/5f/Master_8163_11942.CDS-CDS.lastz.dag.go_c4_D30_g10_A2.aligncoords.gcoords_ct0.w2000.cs1.csoS.nsd.png/488px-Master_8163_11942.CDS-CDS.lastz.dag.go_c4_D30_g10_A2.aligncoords.gcoords_ct0.w2000.cs1.csoS.nsd.png)
![Syntenic dotplot by [SynMap]. X-axis Oryza sativa japonica (Rice); Y-axis Phoenix dactylifera L. (date palm). Results may be regenerated: http://genomevolution.org/r/3qjn](/wiki/images/thumb/8/8a/Master_8163_11942.CDS-CDS.lastz.dag.go_c4_D30_g10_A2.aligncoords.gcoords_ct0.w2000.ass2.cs1.csoS.nsd.png/500px-Master_8163_11942.CDS-CDS.lastz.dag.go_c4_D30_g10_A2.aligncoords.gcoords_ct0.w2000.ass2.cs1.csoS.nsd.png)
![Syntenic dotplot by [SynMap]. X-axis Cannabis sativa (marijuana); Prunus persica (peach). Results may be regenerated: http://genomevolution.org/r/3wz5](/wiki/images/thumb/5/55/Master_12243_8400.genomic-CDS.lastz.dag.go_c4_D20_g10_A2.aligncoords.gcoords_ct0.w2000.cs1.csoS.nsd.png/500px-Master_12243_8400.genomic-CDS.lastz.dag.go_c4_D20_g10_A2.aligncoords.gcoords_ct0.w2000.cs1.csoS.nsd.png)
![Syntenic dotplot by [SynMap]. X-axis Cannabis sativa (marijuana); Prunus persica (peach). Results may be regenerated: http://genomevolution.org/r/3wz3](/wiki/images/thumb/f/f5/Master_12243_8400.genomic-CDS.lastz.dag.go_c4_D20_g10_A2.aligncoords.gcoords_ct0.w500.ass2.cs1.csoS.nsd.png/67px-Master_12243_8400.genomic-CDS.lastz.dag.go_c4_D20_g10_A2.aligncoords.gcoords_ct0.w500.ass2.cs1.csoS.nsd.png)