Genlisea intragenomic analyses: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
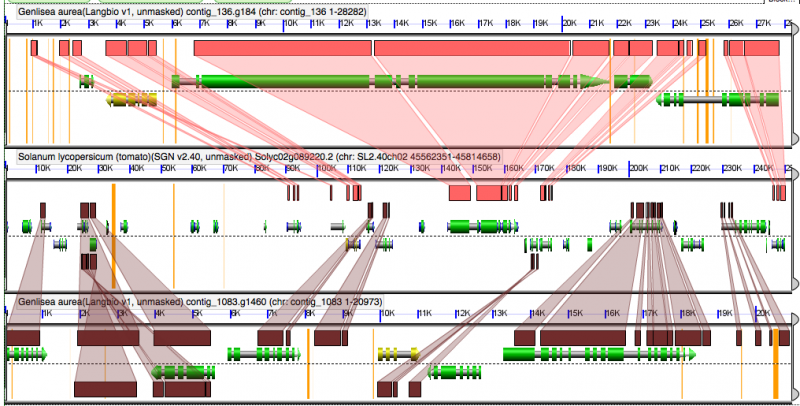
[[File:Screen shot 2012-02-22 at 4.58.00 AM.png|thumb|800px|center|GEvo analysis of syntenic regions of Genlisea (2x) and tomato (1x center). Note that the syntenic genes of Genlisea are intercalated with respect to tomato, but there NO genes are syntenic or homeologously shared between the Genlisea regions. This is strong evidence that Genlisea has a subsequent whole genome duplication followed by extensive fractionation.]] | [[File:Screen shot 2012-02-22 at 4.58.00 AM.png|thumb|800px|center|GEvo analysis of syntenic regions of Genlisea (2x) and tomato (1x center). Note that the syntenic genes of Genlisea are intercalated with respect to tomato, but there NO genes are syntenic or homeologously shared between the Genlisea regions. This is strong evidence that Genlisea has a subsequent whole genome duplication followed by extensive fractionation. Results may be regenerated at: http://genomevolution.org/r/4jf4. Note: there are a few regions of Genlisea that are syntenic and show sequence similarity to annotated genes in tomato. These are probably missed annotations due to unsequenced regions in Genlisea or pseudogenes (degraded former genes).]] | ||
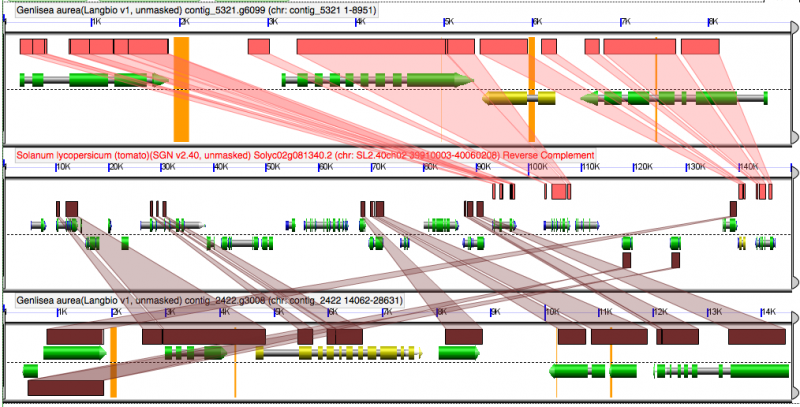
[[File:Screen shot 2012-02-20 at 3.55.26 PM.png|thumb|800px|center|Genlisea regions are NOT intercalated. Slight intercalation might be due to a transposon-like sequence. http://genomevolution.org/r/4j88]] | [[File:Screen shot 2012-02-20 at 3.55.26 PM.png|thumb|800px|center|Genlisea regions are NOT intercalated. Slight intercalation might be due to a transposon-like sequence. http://genomevolution.org/r/4j88]] | ||
Revision as of 12:20, 22 February 2012