Qteller: Difference between revisions
| Line 60: | Line 60: | ||
===Why is qTeller so slow?/Why haven't you included by favorite new dataset yet?=== | ===Why is qTeller so slow?/Why haven't you included by favorite new dataset yet?=== | ||
Please understand I recieve no funding for my work on qTeller. That means it is running on a machine under my desk, not a high powered server, and adding in new data happens only when I have a block of "free time" (as much as it is possible for any graduate student to possess such a thing). That said, if there is a new useful dataset out there which I haven't included I want to hear about it, so don't hesitate to [[ | Please understand I recieve no funding for my work on qTeller. That means it is running on a machine under my desk, not a high powered server, and adding in new data happens only when I have a block of "free time" (as much as it is possible for any graduate student to possess such a thing). That said, if there is a new useful dataset out there which I haven't included I want to hear about it, so don't hesitate to [[User:Jschnable|let me know by e-mail]] if there are new datasets you would like to see included in qTeller's results. | ||
Revision as of 04:46, 25 February 2012
qTeller is a web interface that allows researchers to extract information on genes within an interval and visualize the expression of genes as measured in multiple RNA-seq experiments. By downloading raw sequence data from multiple experiments and piping it all through the same analysis system, it is possible to generate comparable measurements of gene expression from a wide range of tissues, environmental conditions, and mutants. <-- subject always to the caveat that difference in gene expression between different studies may also be explained simply by the fact that plants were grown in different environments.
How Expression Data Is Measured
Reads are aligned to the genome using GSNAP which allows spliced alignments between multiple exons. Format conversion and alignment sorting is carred out using SAMtools and the expression of each annotated gene is quantified using Cufflinks.
Species Studied
Dedicated instances of qTeller are available for maize (25 datasets) and arabidopsis (14 datasets), however the modular nature of qTeller makes it easy to deploy new instances for any species with enough RNA-seq data to make comparative RNA-seq analysis informative.
Example Graphs
Bar Chart
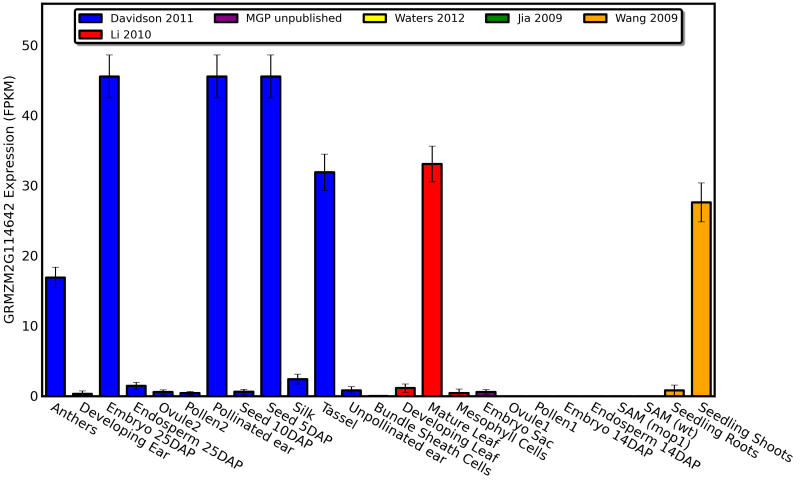
Regenerate this particular analysis by clicking this link
Scatter Plot

FAQ
Where did the name qTeller come from?
qTeller: The website that tells you all about the genes under you QTL. Hey, we had to call it something!
How are the expression values displayed by qTeller calculated?
Raw RNA-seq reads were aligned to the B73 reference genome using GSNAP, and the expression levels of individual genes were quantified using Cufflinks
Why use GSNAP?
As the length of reads generated by Illumina continues to grow, more and more of these reads will span multiple exons. Many of the most popular short-read alignment software packages like Bowtie cannot handle aligning these spliced reads, which means the reads are simply thrown out as unalignable. This is a problem for two reasons:
Throwing out a lot of reads means our measures of gene expression would be less accurate than they otherwise would be. Genes with many small exons (instead of one or a couple of large exons) will disproportionately appear to lose expression as sequencing lengths get longer. GSNAP is one of a handful of aligners that can carry out spliced alignments to a reference genome. A good comparison of some of the alternatives (for example TopHap, MapSplice, and RUM) is presented in this paper: "Comparative Analysis of RNA-Seq Alignment Algorithms and the RNA-Seq Unified Mapper (RUM)."
I have my own set of maize RNA-seq data and I'd like to compare it to all the data in qTeller
Sure! If your dataset is published already and we haven't loaded it into qTeller, send me the link and I will load it in right away. If your dataset isn't published yet, we still might be able to help you out, since it is possible for usto create custom instances of qTeller (even protected by a password if absolutely necessary) which include all these published datasets plus sets of private data.
I have a bunch of RNA-seq data, but it isn't from maize, am I out of luck?
Not necessarily. The modular framework for both qTeller visualizations and the back-end RNA-seq analysis means it's possible to generate an instance of qTeller for practically any species with a reference genome. But it does take a certain critical mass of RNA-seq data before qTeller figures become informative. For an example of what is NOT enough data to really be informative, try generating bar plots or scatter plots using Arabidopsis thaliana gene names. Example.
What is a syntenic ortholog and how are they identified?
When two species are compared, orthologous genes are those descended from the same gene in the most recent common ancestor of those two species. All other similar genes (homologs) are produced by various kinds of duplication (rather than speciation), from the duplication of a individual gene to the duplication of the entire genome of a species. In many cases, distinguishing orthologs from paralogs can be quite difficult, however in the grasses it is possible to do this using a combination of synteny and agregate gene divergence. The syntenic orthologs reported in qTeller were identified using SynMap, part of CoGe.
What is a "classical maize gene"?
Maize geneticists had been identifying and naming mutants in maize for almost a century before the sequencing of the maize genome. A classical maize gene is one that has been the subject of individual study by the maize genetics community and there tends to be a lot more information avaliable about these genes than the average gene in the genome. For more details go classical maize genes here or check out our paper on the subject in PLoS One.
What gave you the idea to create this website?
The idea for this website started when a friend asked me "Can you tell me where the genes under this QTL I'm interested in are expressed?" After the fourth or fifth time I was asked this question (or the related one "I'm really interested in gene X, can you tell me where it is expressed?") I figured I might as well build a web interface so I wouldn't need to look up the expression values for genes in specific regions manually each time. And if I was going to build I website anyway, I figured I would include all the other datasets I use myself when I'm trying to decide which genes I'm most interested in. In maize that means genes with syntenic orthologs in other grasses, and genes which have been previously characterized and studied -- the classical genes of maize genetics -- which are disproprotionately likely to also be genes with conserved syntenic orthologs in other grass species. -James
Why is qTeller so slow?/Why haven't you included by favorite new dataset yet?
Please understand I recieve no funding for my work on qTeller. That means it is running on a machine under my desk, not a high powered server, and adding in new data happens only when I have a block of "free time" (as much as it is possible for any graduate student to possess such a thing). That said, if there is a new useful dataset out there which I haven't included I want to hear about it, so don't hesitate to let me know by e-mail if there are new datasets you would like to see included in qTeller's results.