Jcvi code: Difference between revisions
| Line 112: | Line 112: | ||
Not too bad. | Not too bad. | ||
There is an obvious "decoding" section in the line without a quote (along with some HTML tags) | |||
[[File:Screen Shot 2012-03-22 at 7.54.52 PM.png|600px]] | |||
Revision as of 02:56, 23 March 2012
Background
Rumor has it that there is a code in one of the synthetic genomes by JCVI. Supposedly, this code contains an email address or a URL (or a secret message from Dr. V!).
Synthetic JCVI genomes in CoGe
- synthetic Mycoplasma genitalium strain JCVI-1.0: http://genomevolution.org/CoGe/OrganismView.pl?oid=35986
- synthetic Mycoplasma mycoides JCVI-syn1.0: http://genomevolution.org/CoGe/OrganismView.pl?oid=35385
Methods
- Find closest natural relatives
- Identify syntenic discontinuities (this is where the new JCVI code should reside
- Decode new sequence
- Identify coding scheme
- Probably using natural codon triplet encoding given that:
- 1x4 encoding = 4 letters
- 2x4 encoding = 16 letters
- 3x4 encoding = 64 letters
- Given that there are 20ish natural amino acids, some of the codons will be appropriated for additional letters and symbols
- An example for students of expanded codon encoding (using neighboring codons for additional letters): http://nature.ca/genome/05/051/0511/0511_m205_e.cfm
- Probably using natural codon triplet encoding given that:
- Decode email address
- Identify coding scheme
- Valid email address
Closest natural relatives
| Syntenic dotplot of synthetic Mycoplasma genitalium strain JCVI-1.0 (y-axis) v. Mycoplasma genitalium strain G37 (x-axis) http://genomevolution.org/r/4mx1 | Syntenic dotplot of synthetic Mycoplasma mycoides JCVI-syn1.0 (y-axis) v. Mycoplasma mycoides subsp. capri strain GM12 (x-axis) http://genomevolution.org/r/4mx2 |
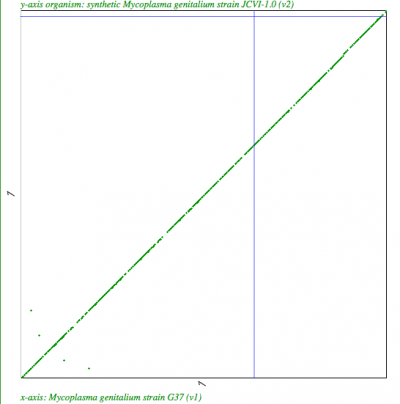
|
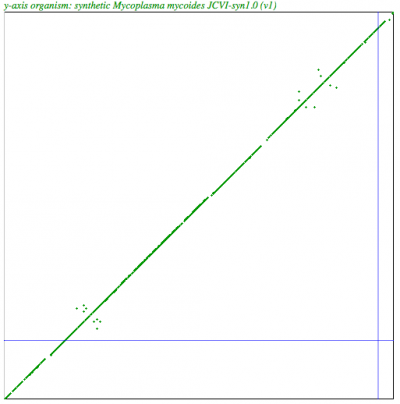
|
GEvo Analyses: high-resolution detection of syntenic discontinuities
Genitalium


- Disrupted WT gene: MG_408 , NP_073081.1 , pmsR
- methionine sulfoxide reductase A
- this stereospecific enzymes reduces the S isomer of methionine sulfoxide while MsrB reduces the R form a fusion protein of this enzyme with MsrB provides protection against oxidative stress in Neisseria gonorrhoeae this stereospecific enzymes reduces the S isomer of methionine sulfoxide while MsrB reduces the R form
- Inserted gene: ABY79711.1 , MGATCC33530_0530
- bifunctional AAC/APH (AAC(6'): 6'-aminoglycoside N-acetyltransferase and APH(2'): 2-aminoglycoside phosphotransferase
- Aminoglycoside antibiotic resistance is largely the result of the production of enzymes that covalently modify the drugs including kinases (Aph) with structural and functional similarity to protein and lipid kinases. One of the most important aminoglycoside resistance enzymes is Aac(6')-Aph(2), a bifunctional enzyme with both aminoglycoside acetyltransferase and kinase activities.
- Extract sequence: http://genomevolution.org/CoGe//SeqView.pl?featid=143193835_1&dsid=63956&chr=1&start=&upstream=-227770&downstream=228068&rc=0&dsgid=15768
- Perform 6-frame translation (using above link)
- search for anything that might match "com/cnm" "org/nrg" "net", "cvi" (jcvi)
- Nothing: could be wrong code
Mycoides
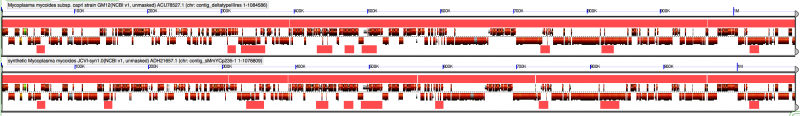
| Region | Notes |
 |
Annotation talks about how a piece of e. coli made it into this area. Not the greener region (GC rich). When that sequence is extracted and blasted against E. coli, get a very high quality hit (95%). http://genomevolution.org/CoGe//CoGeBlast.pl?chr=contig_sMmYCp235-1&upstream=308819&downstream=309545&dsid=59166&rc=0&gstid=1 |
 |
http://genomevolution.org/CoGe//SeqView.pl?dsid=59166&chr=contig_sMmYCp235-1&start=389504&stop=390368&gstid=1. There is an obvious "new thing" that is annotated as a hypothetical protein predicted by glimmer. Gene it replaces is a transposon (which wouldn't be missed). This sequence does not match anything in K12 MG1655. Its sequence, when 6-frame translated, yields one frame that contains several "CVI" amino acid sequences. This sequence hasn't been seen in other aa sequences from this genome. Obvious email/web address did not jump out. |
 |
http://genomevolution.org/CoGe//SeqView.pl?dsid=59166&chr=contig_sMmYCp235-1&start=565568&stop=566608&gstid=1 . Did not match anything in E. coli. No obvious words. Disrupted (removed) WT ABC transporters. |
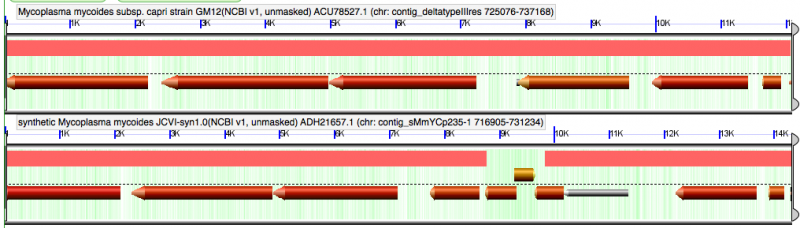 |
http://genomevolution.org/CoGe//SeqView.pl?dsid=59166&chr=contig_sMmYCp235-1&start=725610&stop=726650&gstid=1 . Did not match anything in E. coli. Did contain VCI. Interrupted WT "Type III restriction-modification system is named MmyCI" |
 |
http://genomevolution.org/CoGe//SeqView.pl?dsid=59166&chr=contig_sMmYCp235-1&start=958651&stop=959707&gstid=1 . Did not match anything in E. coli. Did contain VCI. Interrupting a WT transposon. Hit nothing else in NCBI NR database. |
Hints From the Paper and Craig V.
The paper describes four watermark sequences added to the genome. CoGe correctly identified these (along with the carryover piece of E. coli). There is a video and associated articles from Craig describing that the four watermarks contain special phrases. One of which is the URL/email address.
- Paper: http://www.sciencemag.org/content/329/5987/52.abstract
- Sup Data: http://www.sciencemag.org/content/suppl/2010/05/18/science.1190719.DC1/Gibson.SOM.pdf
- http://www.popsci.com/science/article/2010-05/venter-institutes-synthetic-cell-genome-contains-hidden-messages-watermarks
- http://en.wikipedia.org/wiki/Mycoplasma_laboratorium
- JCVI code phrases
- JCVI watermarks (without end sequences)
- Aligned 6-frame translation of watermark 4 shows nothing that matches phrase 4
Decoding part one
The code is something different than using protein translation. Also, there needs to be a way to code special characters, such as "," or ":", etc.
JCVI GEvo analysis of watermarks JCVI notes
Cracking the code (from the repeat sequences)
ATA AAC CTG GGC TAA <s> l i f e TGA ATA TAG GCT ATA TGA TCA TAA CAT ATA t <s> a s <s> t h e y <s> ATA CTG ATA TTT TAG TGC TGC CGT TGA ATA <s> I <s> c a n n o t <s> :)
- Notes:
- spaces all use the same triplet (ATA)
- lower case "i" and upper-case "I" both use the same triplet (CTG): code is case insensitive
First pass output
Not too bad.
There is an obvious "decoding" section in the line without a quote (along with some HTML tags)
![]()