Ancestral angiosperm genomes: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
|||
| Line 18: | Line 18: | ||
[[Eurosid Ancestor to Grape]] | [[Eurosid Ancestor to Grape]] | ||
[[Eurosid Ancestor to Cacao]] | |||
[[Eurosid Ancestor to Peach]] | |||
[[Eurosid Ancestor to Amborella]] | [[Eurosid Ancestor to Amborella]] | ||
Revision as of 18:46, 18 June 2012
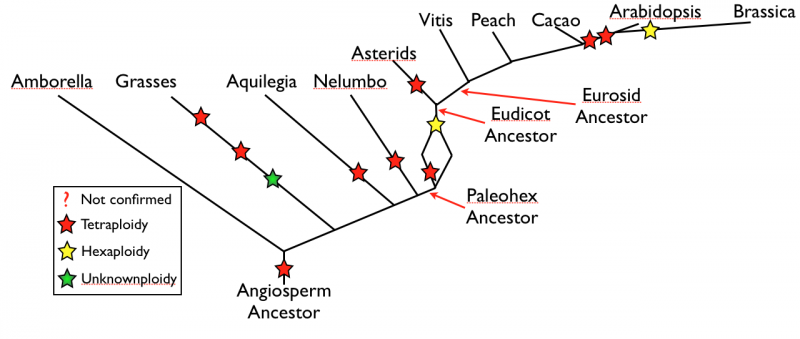
Ancestral Eurosid
- Data generated by Chungfang Zheng
- Procedure for generating ancestral genome:
- Take ancestral ortholog sets from grape/chocolate/peach
- Use the ortholog gene from first grape, then chocolate, then peach (if data is missing)
- Extract CDS sequence from CoGe
- Reverse complement if necessary
- Print sequence
- Pad with 1000 "N"s between coding sequence
Link to ancestral genome: http://genomevolution.org/CoGe/OrganismView.pl?oid=38348