Mimulus v. tomato: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
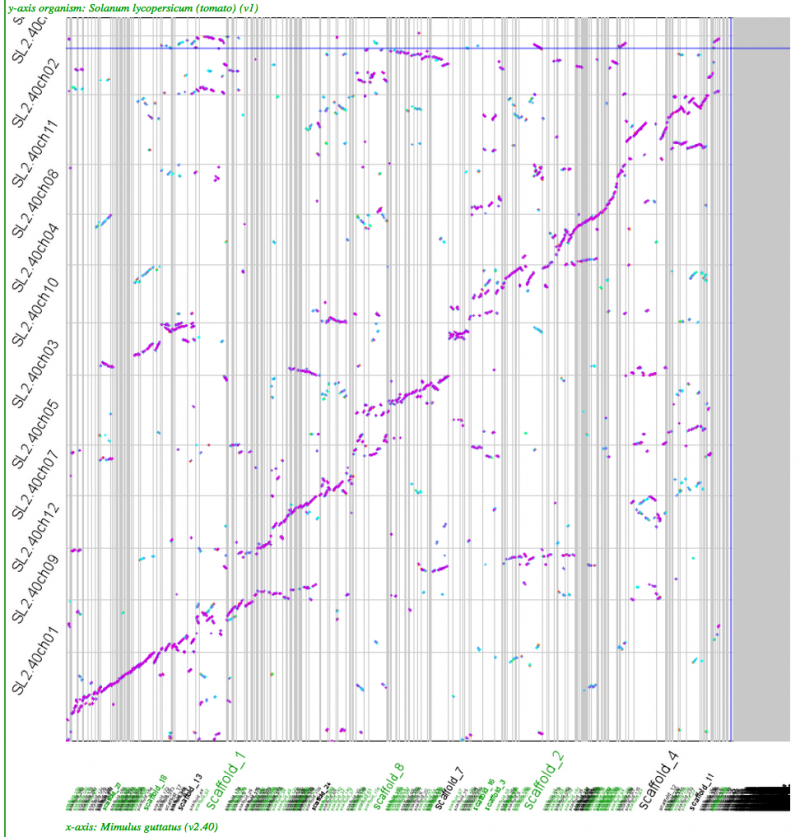
==Whole genome [[syntenic dotplots]] | Note: evaluation of tomato is difficult as its polyploidy is not a string tetraploidy or hexaploidy: [[Tomato genome]] | ||
==Whole genome [[syntenic dotplots]]== | |||
[[File:Screen shot 2012-07-06 at 12.32.07 PM.png|thumb|800px|center|http://genomevolution.org/r/518r]] | [[File:Screen shot 2012-07-06 at 12.32.07 PM.png|thumb|800px|center|http://genomevolution.org/r/518r]] | ||
==High-resolution analysis of syntenic regions with [[GEvo]]== | |||
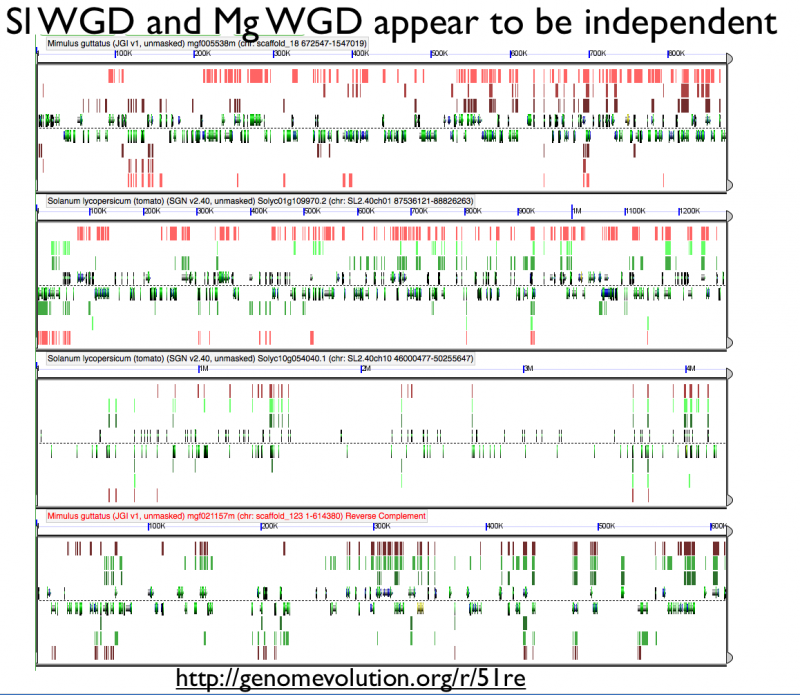
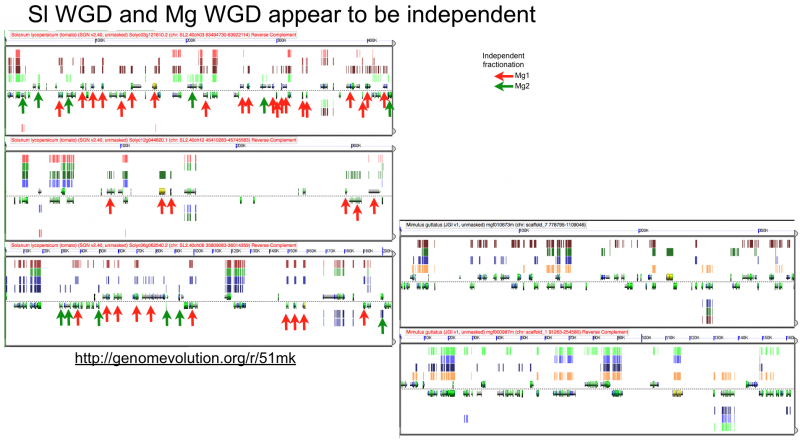
Fractionation appears to be independent among intragenomic syntenic regions | |||
[[File:Screen shot 2012-07-06 at 3.20.46 PM.png|thumb|800px|center| http://genomevolution.org/r/51re]] | [[File:Screen shot 2012-07-06 at 3.20.46 PM.png|thumb|800px|center| http://genomevolution.org/r/51re]] | ||
[[File:Screen shot 2012-07-06 at 3.21.18 PM.png|thumb|800px|center| http://genomevolution.org/r/51mk]] | [[File:Screen shot 2012-07-06 at 3.21.18 PM.png|thumb|800px|center| http://genomevolution.org/r/51mk]] | ||
==Ks histograms of syntenic gene pairs== | |||
Revision as of 14:16, 7 July 2012
Note: evaluation of tomato is difficult as its polyploidy is not a string tetraploidy or hexaploidy: Tomato genome
Whole genome syntenic dotplots
High-resolution analysis of syntenic regions with GEvo
Fractionation appears to be independent among intragenomic syntenic regions