LoadAnnotation: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 13: | Line 13: | ||
===Data File=== | ===Data File=== | ||
You can select and retrieve | Annotations are specified in GFF format (see [[GFF ingestion]] for details). You can select and retrieve the a GFF file from multiple sources: | ||
*The iPlant Data Store | *The iPlant Data Store | ||
*A URL (FTP/HTTP) server | *A URL (FTP/HTTP) server | ||
*Your computer (Upload) | *Your computer (Upload) | ||
Revision as of 18:41, 17 April 2013
LoadAnnotation enables you to load a set of annotation data for a genome in CoGe. The annotation can then be viewed in GenomeView.
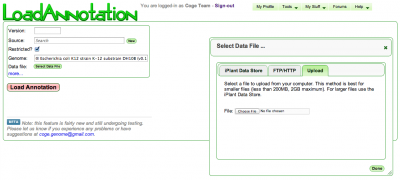
Input
- Version: Version of annotation
- Source: Where is the data from? This could be you, your lab, your university, a sequencing center, your collaborator.
- Restricted: Is this annotation public or restricted to you and your collaborators
- Genome: Select the appropriate genome from CoGe
- Select Data File: Opens a window for specifying the input data file
- Name: Name of annotation data set (optional)
- Description: Description of annotation data set (optional)
Data File
Annotations are specified in GFF format (see GFF ingestion for details). You can select and retrieve the a GFF file from multiple sources:
- The iPlant Data Store
- A URL (FTP/HTTP) server
- Your computer (Upload)