SynMap: Difference between revisions
Jump to navigation
Jump to search
| Line 14: | Line 14: | ||
[[Image:Master_3068_8.CDS-CDS.blastn_geneorder_D20_g10_A5.w2000.ks.png|thumb|right|600px|Syntenic dotplot between ''Arabidopsis thaliana'' and ''Arabidopsis lyrata''. Syntenic gene pairs identified by DAGChainer have been colored based on their synonymous rate change as calculated by CODEML. Results can be regenerated [http://synteny.cnr.berkeley.edu/CoGe/SynMap.pl?dsgid1=3068;dsgid2=8;D=20;g=10;A=5;w=0;b=1;ft1=1;ft2=1;dt=geneorder;ks=1;autogo=1 here]. ]] | [[Image:Master_3068_8.CDS-CDS.blastn_geneorder_D20_g10_A5.w2000.ks.png|thumb|right|600px|Syntenic dotplot between ''Arabidopsis thaliana'' and ''Arabidopsis lyrata''. Syntenic gene pairs identified by DAGChainer have been colored based on their synonymous rate change as calculated by CODEML. Results can be regenerated [http://synteny.cnr.berkeley.edu/CoGe/SynMap.pl?dsgid1=3068;dsgid2=8;D=20;g=10;A=5;w=0;b=1;ft1=1;ft2=1;dt=geneorder;ks=1;autogo=1 here]. ]] | ||
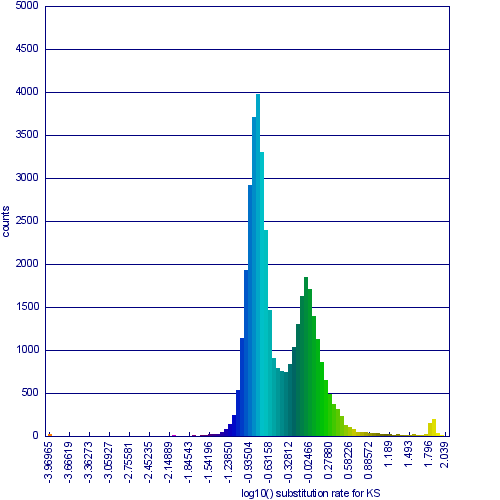
[[Image:Master 3068 8.CDS-CDS.blastn geneorder D20 g10 A5.w2000.ks.hist-1.png|thumb|right|600px|Histogram of synonymous rate change data for syntenic gene pairs between Arabidopsis thaliana and Arabidopsis lyrata.]] | [[Image:Master 3068 8.CDS-CDS.blastn geneorder D20 g10 A5.w2000.ks.hist-1.png|thumb|right|600px|Histogram of synonymous rate change data for syntenic gene pairs between Arabidopsis thaliana and Arabidopsis lyrata. The two obvious peaks in the distribution are from syntenic gene pairs (syntelogs) derived from the speciation of these two taxa and from their shared most recent whole genome duplication event, known as alpha.]] | ||
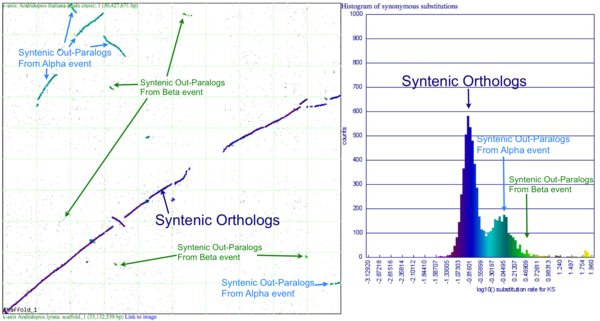
[[Image:SynPlot-Chr1-At-v-Al.png|thumb|right|600px|Syntenic dotplot and synonymous substitution histogram of Chromosome 1 of Arabidopsis thaliana and Arabidopsis lyrata. Syntenic gene pairs were identified by DAGChainer, and colored based on their synonymous substitution rate as calculated by CodeML. Syntenic regions derived from speciation (orthologs) or from the shared whole genome duplication events (alpha and beta) are labeled. These images were generated by SynMap and can be viewed at generated by SynMap at http://toxic.berkeley.edu/CoGe//diags/Arabidopsis_lyrata/Arabidopsis_thaliana_thale_cress/html/3068_8.CDS-CDS.blastn.dag_geneorder_D20_g10_A5.scaffold_1-1.w600.ks.html]] | [[Image:SynPlot-Chr1-At-v-Al.png|thumb|right|600px|Syntenic dotplot and synonymous substitution histogram of Chromosome 1 of Arabidopsis thaliana and Arabidopsis lyrata. Syntenic gene pairs were identified by DAGChainer, and colored based on their synonymous substitution rate as calculated by CodeML. Syntenic regions derived from speciation (orthologs) or from the shared whole genome duplication events (alpha and beta) are labeled. These images were generated by SynMap and can be viewed at generated by SynMap at http://toxic.berkeley.edu/CoGe//diags/Arabidopsis_lyrata/Arabidopsis_thaliana_thale_cress/html/3068_8.CDS-CDS.blastn.dag_geneorder_D20_g10_A5.scaffold_1-1.w600.ks.html]] | ||
Revision as of 16:48, 28 August 2009
Overview
Specifying genomes
DAGChainer options
SynMap options
Calculating and displaying synonymous/non-synonymous (kS, kN) data
Interacting with results
Example Results